Esophagitis ICD 10 A Guide to K20-K21 Coding and Mapping

When documenting esophagitis, the primary ICD-10-CM codes you'll encounter fall within the K20-K21 series. Think of K20 as the category for various other types of esophagitis, while K21 is reserved specifically for gastro-esophageal reflux disease (GERD).
A critical distinction within the GERD classification is whether esophagitis is actually present (K21.0) or absent (K21.9). Getting this right is absolutely essential for accurate clinical documentation, claims processing, and any subsequent data analysis.

Understanding The Core Esophagitis ICD-10 Codes
For anyone working with healthcare data, from clinical researchers to data engineers, a solid grasp of the esophagitis ICD-10 code structure is non-negotiable. The International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) is the bedrock system for diagnosing and categorizing medical conditions, and that includes the various forms of esophageal inflammation.
These codes are the universal language for everything from billing and public health surveillance to building specific patient cohorts for research. A simple misapplication of a code can cascade into denied claims, skewed analytics, and ultimately, flawed study populations. This is why a precise understanding of the K20 and K21 series isn't just about clerical accuracy—it's about maintaining data integrity from the ground up.
Primary Esophagitis ICD-10-CM Codes at a Glance
To start, let's break down the top-level codes that form the foundation for diagnosing esophagitis. The table below provides a quick summary of the primary codes, their official descriptions, and the typical clinical scenarios where you would expect to see them used.
| ICD-10-CM Code | Official Description | Common Clinical Context |
|---|---|---|
| K20 | Other esophagitis | Used for inflammation of the esophagus from causes other than GERD, such as eosinophilic, allergic, or drug-induced esophagitis. |
| K21.0 | Gastro-esophageal reflux disease with esophagitis | Specifically for patients with GERD where endoscopic or clinical evidence of esophageal inflammation is present. |
| K21.9 | Gastro-esophageal reflux disease without esophagitis | Assigned to patients who have symptoms of GERD, but without any observable inflammation of the esophagus. |
This table serves as a high-level reference. Differentiating between these codes correctly is the first step toward building clean, reliable datasets for any kind of analysis or operational process.
Diving Deeper into Key Coding Series
This guide will focus on the two main categories that capture the vast majority of esophagitis diagnoses.
-
K20 Other Esophagitis: This series is your go-to for esophagitis stemming from causes other than acid reflux. This includes conditions like allergic, eosinophilic, or drug-induced inflammation, each with more specific subcodes.
-
K21 Gastro-esophageal reflux disease: This series is dedicated to GERD. Its primary function is to make that crucial distinction between cases presenting with esophagitis (K21.0) and those without (K21.9).
Properly navigating these codes is vital for creating accurate datasets. To understand how these diagnostic codes are structured within larger analytical frameworks, you might find it helpful to read our guide on the OMOP data model. We will explore these codes and their specific applications in much greater detail throughout this reference guide.
The Clinical Weight of K20 and K21 Codes
Getting a handle on the esophagitis icd 10 codes, especially the K20 and K21 series, is more than just a box-ticking exercise. These codes capture a real and growing health problem across the globe. Gastro-esophageal reflux disease (GERD), which is a common reason esophagitis gets diagnosed in the first place, has been on a steep rise for decades. That makes getting the codes right absolutely critical for tracking public health trends and conducting sound clinical research.
This surge really puts a spotlight on why accurate data matters. For data engineers and researchers, telling the difference between various types of esophagitis is the bedrock of building reliable patient cohorts. A single misplaced code can completely throw off epidemiological studies, undermine research results, or lead to misallocating already stretched healthcare resources.
Why Precise Coding Is So Important for Population Health
The climbing rates of GERD and related conditions have a tangible effect on healthcare systems everywhere. The Global Burden of Disease Study revealed a staggering jump in prevalent GERD cases, from 450.76 million in 1990 to 825.60 million in 2021. That's an 83.2% increase. These aren't just numbers; they represent a clear, accelerating trend that data professionals have a responsibility to capture accurately in their datasets. You can dig into the specifics in the full GERD prevalence research.
This trend makes solid vocabulary management a non-negotiable for any team handling healthcare data. When you're building out an ETL pipeline or an analytical model, you have to be confident that a source code like K21.0 is mapped correctly. It’s the only way to pull trustworthy insights from the noise.
Pointers for Data Professionals
For anyone on the front lines of managing clinical data, here are a few practical tips to keep your work with K20 and K21 codes sharp and accurate:
-
Validate Your Mappings: Don't just trust them blindly. Always double-check your ICD-10 to OMOP mappings. A great resource for this is the OMOPHub Concept Lookup, which lets you quickly find standard concepts and see how they relate to one another.
-
Automate What You Can: Manual entry is a recipe for errors. Use APIs to automate vocabulary lookups whenever possible. The OMOPHub SDKs for Python and R can make this process a whole lot smoother.
-
Lean on the Documentation: If you're ever unsure about a concept's relationships or its place in the hierarchy, go to the source. The OMOPHub documentation site has detailed resources that can clear things up.
A Detailed Breakdown of K20 Other Esophagitis Codes
While esophagitis caused by GERD gets a lot of attention, the K20 Other Esophagitis category is where we classify inflammation from a whole different set of triggers. For anyone working with clinical data, getting these non-reflux types right is essential for building accurate patient cohorts and conducting meaningful analysis in platforms like OMOP. This code series is our tool for flagging esophagitis that stems from allergic reactions, chemical ingestion, or even medication side effects.
Getting the specific esophagitis icd 10 codes right prevents the kind of misclassification that can seriously skew research findings or cause billing headaches. The first step to achieving that high-fidelity data capture is really digging into the details of each subcode.
Unpacking the K20 Subcodes
The K20 category isn't a monolith; it's broken down into more granular diagnoses, each pointing to a unique cause. These are the main subcodes you'll be working with:
- K20.0 Allergic and eosinophilic esophagitis: This is your code when the inflammation is driven by an allergic process. The classic sign is a high concentration of eosinophils found in esophageal tissue biopsies.
- K20.8 Other esophagitis: Think of this as the "specified but not otherwise classified" bucket. It's the correct choice for clear-cut cases like drug-induced esophagitis or corrosive damage from swallowing chemicals.
- K20.9 Esophagitis, unspecified: This code is used when the clinician confirms esophagitis is present, but the underlying cause hasn't been pinned down or documented yet. It’s a necessary, but less useful, code for data analysis.
The Rise of Eosinophilic Esophagitis (K20.0)
Eosinophilic esophagitis (EoE), which falls under K20.0, has become much more commonly diagnosed in recent years. This makes its correct identification in datasets more critical than ever. A meta-analysis pooling 29 different studies revealed a global prevalence of 34.4 cases per 100,000 people—a number that underscores its growing clinical importance. For ETL developers and data mappers, getting this condition mapped correctly is non-negotiable for robust data harmonization. You can dive into the full research on EoE prevalence to get a better sense of its epidemiological footprint.
One of the most common pitfalls in data processing is failing to properly differentiate K20.0 from standard reflux esophagitis. It's a critical distinction. Lumping patients with an autoimmune-driven condition into the same cohort as those with a digestive disorder can completely compromise the integrity of clinical research.
For those of you building and maintaining data pipelines, here are a few practical tips to keep your data clean:
- Prioritize Specificity: Always go for the most specific code available. If the clinical notes mention an allergic component or eosinophil counts, K20.0 is far more accurate than the generic K20.9.
- Cross-Reference Your Sources: Don't just rely on the coded diagnosis. Use clinical notes and, most importantly, pathology reports to confirm the etiology. This is the best way to distinguish an allergic cause from a reflux-related one.
- Lean on Your Vocabulary Tools: Resources like the OMOPHub Concept Lookup are invaluable for exploring the relationships between ICD-10 codes and standard terminologies like SNOMED CT. This helps ensure your mapping logic is sound.
A Guide to K21 Gastro-Esophageal Reflux Disease Codes
The K21 series is the home for Gastro-Esophageal Reflux Disease (GERD), a diagnosis you'll encounter constantly in gastroenterology data. For anyone working with this data, the most critical thing to understand about this category is its primary split: is there esophageal inflammation, or not?
This single detail divides the category into two foundational codes, and the distinction has massive implications for everything from building patient cohorts to billing and clinical analysis. If you misassign these codes, you risk serious data integrity problems, like polluting a control group of patients with only reflux symptoms by mixing in those with active esophageal damage. Getting this right isn't a minor detail—it's fundamental to accurate research.

Distinguishing K21.0 from K21.9
Grasping the difference between the two main K21 codes is essential for anyone handling clinical data. The decision always comes down to documented evidence of esophagitis.
- K21.0 Gastro-esophageal reflux disease with esophagitis: Use this code when a patient has GERD and there’s clear clinical or endoscopic evidence of inflammation in the esophagus. This is the more severe diagnosis, pointing to actual tissue damage.
- K21.9 Gastro-esophageal reflux disease without esophagitis: This is the correct code for patients with classic GERD symptoms—heartburn, acid regurgitation—but no visible inflammation or esophageal damage on examination.
The ICD-10 framework places the esophagitis icd 10 codes (K20-K21) right at the intersection of GERD and other inflammatory issues. Recent temporal data shows just how much the burden of these conditions has grown. GERD incident cases shot up 80.1%, from 180.0 million in 1990 to 324.1 million in 2021, with incidence rates peaking in the 30–39 age group. You can dig deeper into these epidemiological trends and their impact. For analytics teams standardizing data in an OMOP common data model, this kind of demographic context is invaluable.
Practical Tips for ETL Developers and Data Analysts
To keep your data clean when processing K21 codes, stick to these best practices:
- Scrutinize Endoscopy Reports: The gold standard for telling K21.0 apart from K21.9 is to cross-reference with endoscopy reports. Your logic should hunt for keywords like "inflammation," "erythema," or "erosions" to confirm esophagitis.
- Don't Code from Symptoms Alone: Avoid assigning a K21 code based on symptoms in isolation. If a GERD diagnosis isn't explicitly confirmed, a symptom code like R12 (Heartburn) might be a more accurate choice for the raw data.
- Watch for Co-Occurring Conditions: Always be on the lookout for related diagnoses. Hiatal hernia (K44.9) is a frequent companion to GERD and will likely need to be coded separately to capture the full clinical picture.
Mapping Esophagitis Codes to OMOP Standard Concepts
When you're working with raw clinical data, one of the first and most critical steps for any serious analysis is standardization. Taking source codes like the esophagitis ICD-10 codes K20 and K21 and transforming them into a standard format is fundamental. Within the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), this means mapping source codes from vocabularies like ICD-10-CM to standard concepts, which almost always come from SNOMED CT.
This isn't just a technical data-shuffling exercise. It's about achieving genuine data harmonization, making it possible to compare apples to apples across entirely different healthcare systems. By translating billing codes into a much richer clinical terminology, you establish a semantic consistency that unlocks far more powerful analytics.
Why Standard Vocabulary Mapping Matters
Think about it this way: ICD-10 is a classification system built primarily for billing and epidemiology. SNOMED CT, on the other hand, is a comprehensive clinical ontology designed for detailed, granular documentation. This hierarchical structure lets you build more precise patient cohorts and run analyses based on specific clinical meanings instead of overly broad billing categories.
The whole point is to create a unified dataset for analysis. A patient diagnosed with "Gastro-esophageal reflux disease with esophagitis" might get coded as K21.0 at one hospital, while another facility might use a slightly different description in their records. Mapping both of these instances to a single, standard SNOMED CT concept ensures they're recognized as the same clinical event. This is absolutely essential for multi-site studies or any effort to generate reliable evidence from diverse data sources.
Building this mapping logic is a cornerstone of any reliable ETL pipeline. It's such a foundational concept that we've written an entire guide on the principles of semantic mapping in healthcare data.
A common pitfall is to treat ICD-10 codes as if they have the same clinical depth as a standard terminology. For instance, 'K20.9, Esophagitis, unspecified' is perfectly fine for billing, but it's too vague for most advanced research. Mapping it to a standard concept forces your data team to consciously decide how to handle that ambiguity.
Finding the Right Standard Concepts
Pinpointing the correct standard concept for a given source code is where the real work begins. You could try to look up these mappings manually, but that approach is slow, tedious, and highly susceptible to error.
A much better way is to use a purpose-built tool. The OMOPHub Concept Lookup tool, available at https://omophub.com/tools/concept-lookup, is designed for exactly this. You can plug in a source code like 'K21.0' and immediately see its corresponding standard SNOMED CT concept. More importantly, you can explore its relationships within the OMOP vocabulary hierarchy, which is crucial for building accurate and consistent ETL logic.
You can find more detailed guides on using these kinds of tools in the official documentation on vocabulary exploration at OMOPHub.
To give you a practical sense of what this looks like, here are some common mappings for esophagitis codes.
Example Mappings from ICD-10-CM to OMOP Standard Concepts (SNOMED CT)
This table shows common esophagitis ICD-10-CM codes and their corresponding standard concept mappings within the OMOP CDM, typically to SNOMED CT concepts.
| Source Code (ICD-10-CM) | Source Description | Standard Concept ID (OMOP) | Standard Concept Name (SNOMED CT) | Concept Class |
|---|---|---|---|---|
| K20.9 | Esophagitis, unspecified | 4249969 | Esophagitis | Clinical Finding |
| K21.0 | Gastro-esophageal reflux disease with esophagitis | 432586 | Gastroesophageal reflux with esophagitis | Clinical Finding |
| K21.9 | Gastro-esophageal reflux disease without esophagitis | 439845 | Gastroesophageal reflux disease | Clinical Finding |
| K20.8 | Other esophagitis | 40480336 | Other specified esophagitis | Clinical Finding |
As you can see, the mapping process connects a code used for one purpose (billing) to a concept designed for deep clinical representation. This translation is what makes large-scale, federated network research possible.
Automating Vocabulary Lookups with the OMOPHub API
Interactive tools like the OMOPHub Concept Lookup are fantastic for exploring a few codes here and there, but when you're dealing with real-world data volumes, manual lookups just don't scale. To build serious ETL pipelines, create rich feature sets for machine learning, or run dynamic cohort queries, you need to automate. This is where programmatic access through an API really shines.
With the OMOPHub API, you can programmatically map an esophagitis ICD-10 code directly to its standard OMOP concept. This approach sidesteps the risk of manual error, enforces consistency across your entire dataset, and massively speeds up your data processing jobs. Instead of a person looking up codes one by one, your scripts can handle thousands of them in a flash.
The typical data flow is pretty straightforward: a source code from ICD-10 gets processed through an ETL pipeline, loaded into the OMOP CDM, and standardized to a SNOMED CT concept.
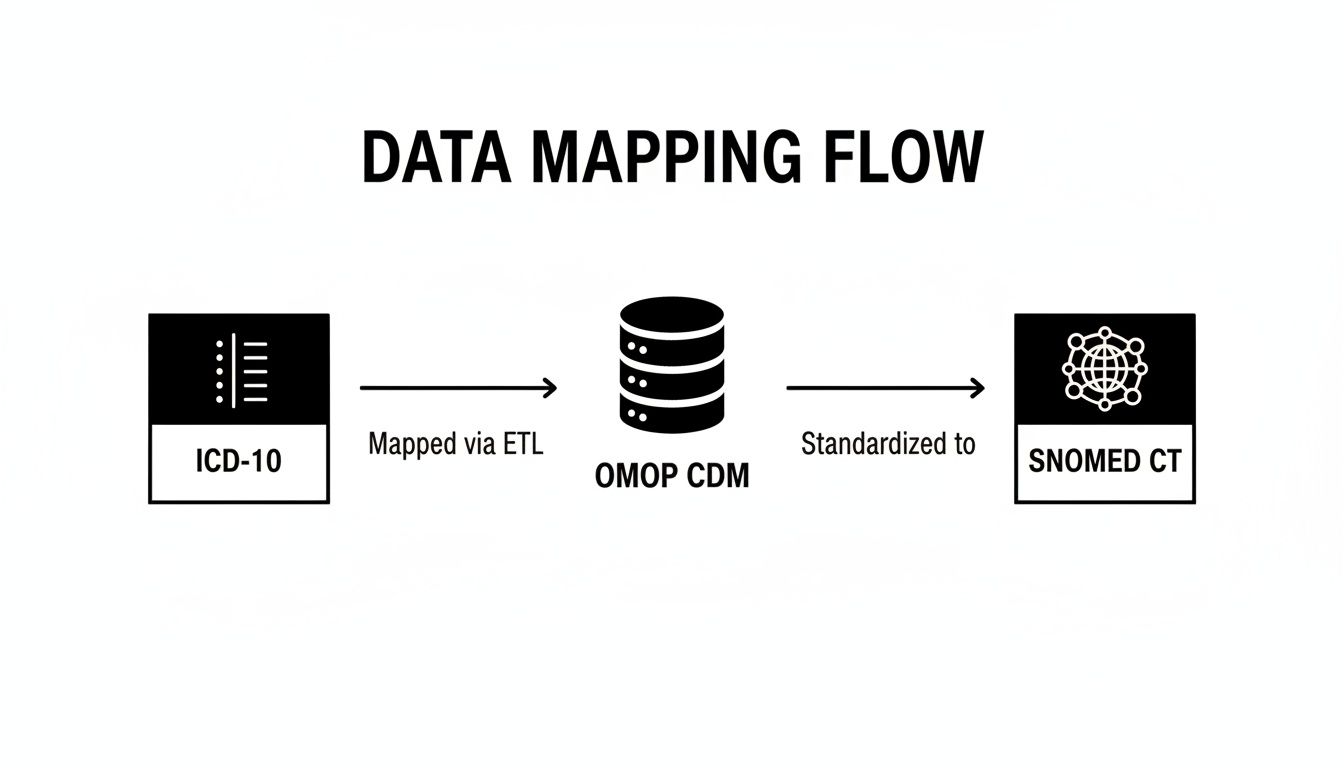
This diagram really underscores how crucial a structured vocabulary mapping process is. It's the key to achieving true data interoperability and getting your information ready for any kind of large-scale analysis.
Getting Started with Programmatic Lookups
OMOPHub offers production-ready SDKs for both Python and R, which makes it incredibly easy to drop vocabulary functions right into your existing analytics code. These toolkits handle the messy parts of direct API calls, giving you simple functions to find concepts, walk their relationships, and validate your mappings. You can find them on GitHub here: https://github.com/OMOPHub/omophub-python and https://github.com/OMOPHub/omophub-R.
For instance, a simple Python script can take an ICD-10 code like K21.0 and pull its corresponding standard concept ID from SNOMED CT.
# Example using the omophub-python SDK
from omophub.client import Client
# Initialize the client with your API key
client = Client(api_key="YOUR_API_KEY")
# Look up an ICD-10-CM code for esophagitis
# This corresponds to the official documentation example
response = client.vocabulary.lookup_source_code(
source_vocabulary_id='ICD10CM',
source_code='K21.0',
)
# Get the standard concept
standard_concept = response['standard_concept']
print(f"ICD-10 Code: K21.0")
print(f"Standard Concept ID: {standard_concept['concept_id']}")
print(f"Standard Concept Name: {standard_concept['concept_name']}")
Tips for Effective API Integration
To get the most out of programmatic lookups, it helps to follow a few battle-tested practices.
- Batch Your Requests: When you're churning through large datasets, try to bundle multiple code lookups into a single API request if the API supports it. This cuts down on network overhead and is much more efficient.
- Handle Non-Standard Mappings: Be prepared for source codes that don't have a clean, one-to-one standard mapping. Your code should have logic to handle these cases, perhaps by using the relationship endpoints to find a suitable parent concept. For a deeper dive, check out the official API documentation on OMOPHub.
- Cache Frequent Lookups: If you find yourself querying the same codes over and over, set up a simple local cache. This can dramatically speed up ETL jobs and save you from making redundant API calls.
For anyone working with different coding systems, our guide on ICD-10 to ICD-9 conversion has some useful tips for managing vocabulary versions. And if you want to take automation a step further, understanding how NLU in healthcare transforms operational efficiency can provide some powerful ideas. This level of programmatic control is what allows a data team to build scalable, accurate, and truly maintainable healthcare data solutions.
Common Questions About Esophagitis Coding
When you're working with esophagitis codes, a few questions tend to pop up again and again, especially for data teams. Let's walk through some of the most common sticking points to clear up any confusion and help you get your data pipelines right.
The K20.9 vs. K21.0 Puzzle
One of the most frequent mix-ups I see is between K20.9 and K21.0. While both point to inflammation in the esophagus, they tell very different clinical stories.
- K20.9 (Esophagitis, unspecified) is your go-to when the record confirms inflammation, but the clinician hasn't documented a specific cause. Think of it as a placeholder—we know there's a problem, but the "why" is still missing.
- K21.0 (Gastro-esophageal reflux disease with esophagitis) is much more precise. You should only use this code when the documentation explicitly links the esophagitis to GERD. It’s not just inflammation; it’s inflammation caused by reflux.
Getting this distinction right is absolutely critical for accurate cohort building. If you're running analytics or conducting research, mistaking one for the other can completely skew your patient populations and lead to flawed conclusions.
Navigating Mapping Gaps in OMOP
So, what do you do when an ICD-10 code doesn't have a clean, one-to-one mapping in the OMOP Common Data Model? This is a classic ETL headache. Not every source code has a perfect dance partner in the standard terminologies.
When you hit this wall, the standard practice is often to map the code to a broader parent concept in the SNOMED CT hierarchy. Sometimes, this requires a bit of clinical judgment to pick the best-fit concept that doesn't lose the original diagnostic intent. Tools like the OMOPHub Concept Lookup are built for exactly this scenario, letting you explore the concept relationships to make a sound decision. You can check out the tool here: https://omophub.com/tools/concept-lookup.
A key principle here is that harmonization is the ultimate goal. It's almost always better to map to a slightly more general standard concept than to leave the source code unmapped. An unmapped code is a dead end for analysis, but a well-chosen standard concept keeps that data point in play for larger, multi-site studies.
This process of mapping to a standard terminology like SNOMED CT is what allows for powerful, large-scale research. It’s how we align data from countless different sources, sharpen our cohort definitions, and ultimately produce more reliable evidence.
At OMOPHub, we create developer-first tools to take the pain out of managing vocabulary databases. We help your team build scalable, compliant, and accurate healthcare data solutions. Find out more and get started at https://omophub.com.


