A Developer Guide to ICD10 to ICD9 Conversion with OMOP

Navigating the world of healthcare data often means looking backward as much as forward. For data engineers and clinical researchers, converting modern ICD-10 codes back to the older ICD-9 system is a frequent, yet surprisingly complex, task. This "reverse mapping" is a critical step for any project that needs to bridge the pre- and post-2015 data divide, especially in long-term outcomes research or historical trend analysis.
The root of the problem isn't just a simple code lookup; it's a fundamental mismatch in detail. Modern ICD-10 codes are surgically precise, while their ICD-9 predecessors were often much broader. This means a single, highly specific ICD-10 diagnosis can map back to a more general ICD-9 equivalent, or worse, have no direct match at all.
The Legacy Challenge of ICD10 to ICD9 Conversion

The switch from ICD-9 to ICD-10 wasn't just an update; it was a complete overhaul of the medical coding landscape. This transition dramatically expanded the number and granularity of codes available to clinicians and researchers.
To put it in perspective, the number of diagnosis codes exploded from roughly 14,000 in ICD-9-CM to over 70,000 in ICD-10-CM. This five-fold increase was fueled by a more detailed, alphanumeric structure designed for greater specificity. For a deeper dive into how this shift impacted data systems, this in-depth analysis of ICD coding systems is an excellent resource.
Let's look at the core differences that create these conversion headaches.
Key Differences Between ICD-10 and ICD-9
The table below breaks down the structural and content changes that are the root cause of the mapping complexity.
| Attribute | ICD-10-CM | ICD-9-CM |
|---|---|---|
| Number of Codes | 70,000+ | 14,000+ |
| Code Structure | 3-7 alphanumeric characters | 3-5 numeric characters |
| Specificity | High (includes laterality, encounter type) | Low (general diagnoses) |
| Category Expansion | 21 chapters | 17 chapters |
| Example | S72.031A: Displaced pincer-type fracture of the head of the right femur, initial encounter | 820.03: Fracture of transcervical section of neck of femur |
As you can see, the leap in detail from ICD-9 to ICD-10 is significant. This inherent "granularity gap" is what we must bridge.
Bridging the Granularity Gap
This expansion means that a very specific ICD-10 code—like S72.031A for a displaced pincer-type fracture of the right femur's head during an initial encounter—often collapses into a much broader ICD-9 code like 820.03, "Fracture of transcervical section of neck of femur." The loss of detail about laterality (right vs. left) and encounter type is simply unavoidable.
This discrepancy in specificity is the central problem. A successful conversion strategy must account for these "one-to-many" or "approximate" mappings in a consistent and clinically sound manner.
This is exactly where a robust vocabulary model becomes non-negotiable. For teams working on projects like health economics and outcomes research (HEOR), just grabbing a simple crosswalk file off the internet won't cut it. A reliable process demands a sophisticated understanding of the relationships between medical concepts to maintain data integrity.
Without this, you're not just losing detail—you're actively introducing bias into your datasets, which can invalidate your entire analysis. This is why frameworks like the OMOP Common Data Model are so valuable; they provide the structured, systematic approach needed to handle these complexities correctly.
2. Choosing Your Mapping Strategy: GEMs vs. OHDSI Vocabularies
Picking the right mapping strategy is probably the most critical decision you'll make when converting ICD-10 back to ICD-9. This choice will ripple through your entire project, affecting everything from the clinical accuracy of your data to the long-term maintainability of your ETL pipelines.
Essentially, you have two main paths: the legacy General Equivalence Mappings (GEMs) or the more modern, relationship-driven vocabularies from OHDSI.
The Old Guard: Why GEMs Fall Short
For a long time, GEMs were the only game in town. Developed by the Centers for Medicare & Medicaid Services (CMS), they were a necessary tool to help the industry bridge the gap from ICD-9 to ICD-10. But they were built for a one-time transition, and it shows. They’re basically just static crosswalk files.
The biggest headache with GEMs is their complex flagging system. A flag might tell you a match is approximate, or that one code needs to map to several others, or that you need a combination of codes to create an equivalent meaning. This was a noble attempt at precision, but it makes automation a nightmare. You practically need a Rosetta Stone and a team of clinical coding experts just to interpret the flags correctly.
Their static nature is another killer. Medical knowledge isn't static. For instance, the FY 2026 ICD-10-CM update brought in 487 new diagnosis codes. GEMs don't automatically account for this. This leaves your data team stuck in a frustrating loop of hunting down, validating, and manually implementing new files, which adds a ton of risk and overhead to your process.
The fundamental problem with GEMs is that they're a snapshot in time. They act as a simple translation dictionary, not the living, breathing semantic network that modern data analytics and harmonization projects really need.
A Better Way: The OHDSI Vocabularies
This is where the OHDSI approach really shines. It doesn't just see codes; it sees an interconnected web of clinical concepts. Instead of a flat crosswalk, the OHDSI Standardized Vocabularies use a sophisticated model of relationships between concepts.
When you perform an ICD-10 to ICD-9 conversion using this model—whether through the ATHENA portal or an API like OMOPHub—you're not just doing a lookup. You're following a clearly defined relationship, most often a 'Maps to' link.
This is a fundamentally better way to work for a few key reasons:
- It's Alive and Maintained: The OHDSI community actively curates and updates the vocabularies. New codes get added, relationships get refined, and you get the benefit without having to manage it all yourself.
- No More Guesswork: A 'Maps to' relationship is a clear, machine-readable statement of equivalence. It removes all the ambiguity you get from trying to decipher the GEMs' flag system.
- Built for a Standard: Mapping with OHDSI aligns your data with a global standard—the OMOP Common Data Model. This is a massive win for interoperability and makes it possible to participate in larger network research studies.
Head-to-Head Comparison
The OHDSI model is simply a more robust and scalable foundation for any serious data conversion work. Let's break it down:
| Feature | General Equivalence Mappings (GEMs) | OHDSI Vocabularies (via OMOPHub) |
|---|---|---|
| Structure | Static, flat crosswalk files | Dynamic, relational concept database |
| Updates | Manual; you have to find and load new files | Centrally managed with official releases |
| Ambiguity | Uses a complex, multi-part flag system | Uses clear, explicit relationship types |
| Integration | Standalone; requires building custom logic | Native to the OMOP CDM; easily accessible via API |
For anyone building a modern, resilient ETL pipeline, the choice is pretty clear. While it's good to know the history of GEMs, the OHDSI vocabulary model offers a far more stable and future-proof path for accurate, automated ICD-10 to ICD-9 conversion.
Tools like the OMOPHub API make this even easier by giving you direct, programmatic access to these carefully curated relationships. If you want to see how it works, the OMOPHub API documentation is a great place to start digging into the concepts.
Putting Theory Into Practice: Automating Conversion with the OMOPHub API
Alright, we've covered the mapping strategies. Now for the real work: getting this conversion automated inside a scalable data pipeline. This is where a specialized tool like the OMOPHub API comes into play, giving you direct, programmatic access to the full OHDSI ATHENA vocabularies.
The whole point is to ditch the manual lookups and clunky static files. Instead, you make dynamic API calls to a centrally managed, always-current vocabulary database. It's a massive time-saver for developers, but more importantly, it ensures your mappings don't become stale as soon as a new vocabulary version is released.
This diagram lays out the two main paths you can take. It clearly contrasts the old-school, file-based GEMs approach with the more modern, relationship-driven OHDSI model.
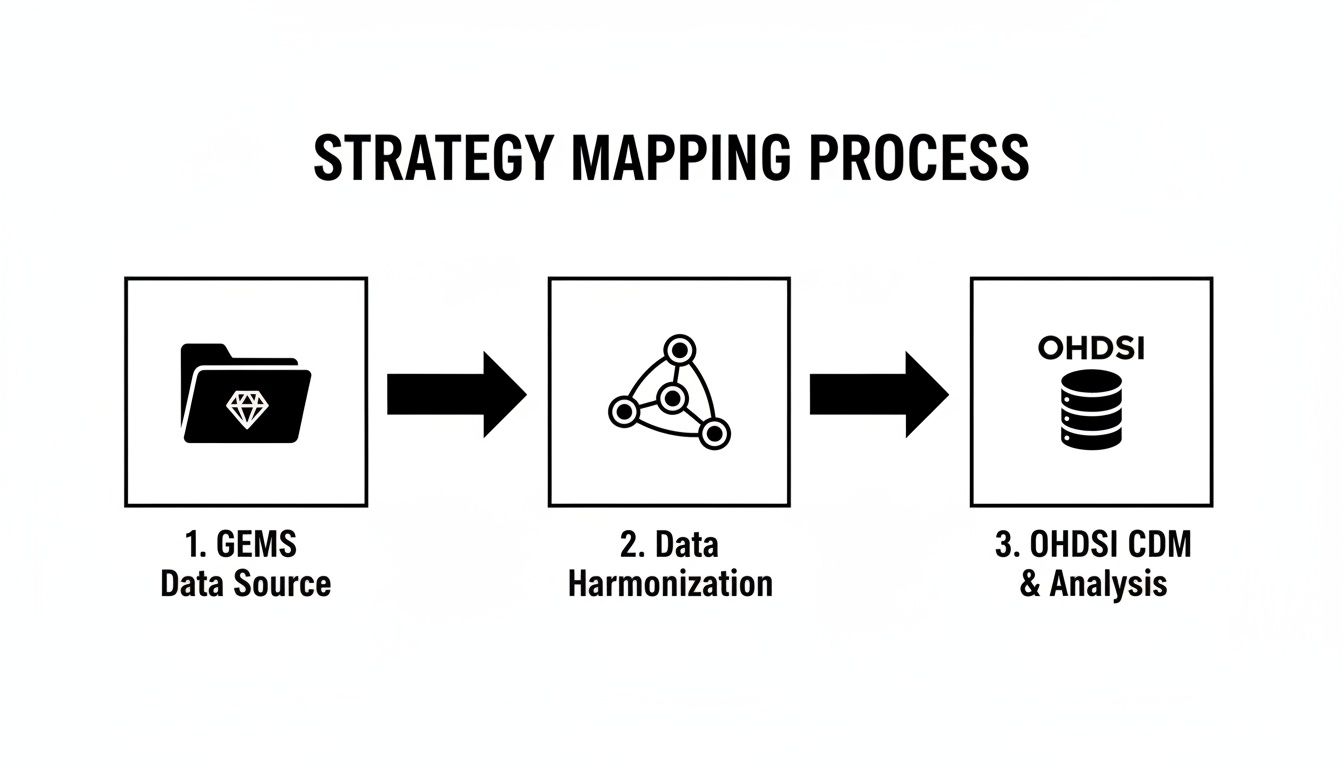
As you can see, the OHDSI route gives you a much cleaner, more integrated workflow. You're using a standardized vocabulary to harmonize your data straight into the OMOP Common Data Model, which is exactly where you want to be.
Getting Started with the OMOPHub SDKs
First things first, you'll need to grab the right OMOPHub SDK for your tech stack. They offer SDKs for Python and R, which covers the vast majority of environments used for data engineering and health analytics today.
You can find the official SDKs right on GitHub:
Installation is as simple as it gets. If you're working in Python, for example, it's just a standard pip install.
pip install omophub
With the SDK installed, you just need to instantiate a client with your API key. This client is your portal to the entire ATHENA vocabulary. No more hosting and maintaining a massive local database—just pure, simple access to search for concepts and trace their relationships.
A Practical Python Example
Let's walk through a real-world scenario. Say you have an ICD-10-CM code and need to find its ICD-9-CM equivalent. Our goal is to take that code, locate its corresponding concept within the OHDSI vocabulary, and then follow its relationships to find what it 'Maps to'.
First, we'll set up the client.
import os
from omophub.client import OMOPHubClient
# Tip: Store your API key as an environment variable for security
api_key = os.environ.get("OMOPHUB_API_KEY")
client = OMOPHubClient(api_key=api_key)
Next, let's find the concept for ICD-10-CM code I21.3 (ST elevation myocardial infarction of unspecified site). We'll use the search endpoint for this. You can dig into all its parameters in the OMOPHub API documentation later.
# Search for the ICD-10-CM concept
# We specify the vocabulary_id and concept_code to get an exact match
search_results = client.concepts.search(
vocabulary_id=["ICD10CM"],
concept_code=["I21.3"],
limit=1
)
# Extract the concept ID from the response
if search_results and len(search_results) > 0:
icd10_concept = search_results[0]
icd10_concept_id = icd10_concept.concept_id
print(f"Found ICD-10 Concept ID: {icd10_concept_id}")
Perfect. Now that we have the unique concept_id for our ICD-10 code, we can easily pull its relationships.
# Get relationships for the concept
relationships = client.concepts.get_relationships(concept_id=icd10_concept_id)
# Filter for 'Maps to' relationships targeting ICD-9-CM
for rel in relationships:
if rel.relationship_id == "Maps to" and rel.concept.vocabulary_id == "ICD9CM":
print(f"Maps to ICD-9-CM code: {rel.concept.concept_code} - {rel.concept.concept_name}")
Pro Tip: When you're building this into your ETL scripts, don't forget robust error handling. Always check that the API response actually contains results before you try to access them. It's a simple step that will keep your script from crashing if a source code is invalid or just doesn't have a direct mapping.
This kind of programmatic approach is a game-changer for EHR integrators and data engineering teams. They're constantly dealing with mapping demands, and legacy tools like GEMs just don't cut it for complex cases, as documented in studies on global ICD-10 adoption challenges. OMOPHub gives you a reliable method for traversing these relationships, and its global edge caching means you get fast responses every time. Your pipelines become not just more accurate, but also resilient to future vocabulary updates. Keep an eye on the OMOPHub changelog to see how the SDKs continue to evolve.
Handling Ambiguous and One-to-Many Mappings
The real challenge in any automated icd10 to icd9 conversion isn't the easy one-to-one matches. The true test is how your pipeline handles the messy, real-world scenarios—specifically, the ambiguous and one-to-many mappings that are unavoidable when you’re translating from a granular system like ICD-10 back to the broader categories of ICD-9.

A classic example is when a single, very specific ICD-10 code could reasonably map to several different ICD-9 codes. Think of an ICD-10 code that specifies laterality (left vs. right). If the source data didn’t capture that detail, you might get two potential ICD-9 matches—one for the left side, one for the right. Your ETL logic has to be smart enough to navigate this without manual intervention.
Strategies for Resolving Ambiguity
If you don't have a plan, these situations will inject a ton of noise and inconsistency into your final dataset. The solution is to define a clear set of business rules that your pipeline can execute reliably every single time it hits one of these forks in the road.
Here are a few battle-tested approaches I’ve seen work well:
- Pick a "Primary" Mapping: Some vocabularies will actually flag one mapping as the preferred or primary choice. It's often the simplest solution: just code your logic to default to that primary mapping and move on.
- Establish a Clinical Hierarchy: Work with your clinical experts to create a set of rules. A common one is to always prioritize a more specific ICD-9 code over a vague "unspecified" option when both are presented as possibilities.
- Log Everything for Review: This is my preferred approach for its transparency. The pipeline loads the most likely mapping into the main table but logs all potential mappings into a separate audit or QA table. This lets the ETL process complete without getting stuck, while flagging complex cases for a human to review later.
The most important thing is to make a conscious, documented decision. An inconsistent, ad-hoc approach to ambiguity is far more dangerous to your data integrity than picking a single, well-understood strategy—even if that strategy sometimes involves a small, calculated loss of information.
Identifying Multiple Mappings in Code
The first step is teaching your code to recognize when the OMOPHub API returns more than one valid 'Maps to' relationship. This means your script needs to parse the API response, loop through the results, and build a list of all potential ICD-9 targets for a given ICD-10 code.
This Python snippet shows exactly how to do that. It takes a concept ID, fetches all its relationships, and then isolates just the ones that map to the ICD-9-CM vocabulary.
import os
from omophub.client import OMOPHubClient
# Tip: Encapsulate mapping logic in a function for reusability.
def find_icd9_mappings(client, icd10_code):
"""
Finds ICD-9-CM mappings for a given ICD-10-CM code.
"""
try:
# 1. Find the source concept
source_concepts = client.concepts.search(
vocabulary_id=["ICD10CM"],
concept_code=[icd10_code],
limit=1
)
if not source_concepts:
return f"No concept found for {icd10_code}"
source_concept_id = source_concepts[0].concept_id
# 2. Get its relationships
relationships = client.concepts.get_relationships(concept_id=source_concept_id)
# 3. Filter for 'Maps to' relationships to ICD9CM
icd9_mappings = [
rel.concept for rel in relationships
if rel.relationship_id == "Maps to" and rel.concept.vocabulary_id == "ICD9CM"
]
# 4. Return the results based on business logic
if len(icd9_mappings) == 0:
return "No direct ICD-9 mapping found."
elif len(icd9_mappings) == 1:
return f"One-to-one mapping: {icd9_mappings[0].concept_code} - {icd9_mappings[0].concept_name}"
else:
output = "One-to-many mapping. Potential codes:\n"
for m in icd9_mappings:
output += f"- {m.concept_code}: {m.concept_name}\n"
return output.strip()
except Exception as e:
return f"An error occurred: {e}"
# Example Usage:
api_key = os.environ.get("OMOPHUB_API_KEY")
client = OMOPHubClient(api_key=api_key)
print(find_icd9_mappings(client, "I21.3")) # An example of a one-to-one
print(find_icd9_mappings(client, "M19.011")) # An example that may have multiple mappings
This simple check—just the length of the icd9_mappings list—is the cornerstone of a robust handling process. It allows you to branch your ETL script into different logic paths, ensuring every icd10 to icd9 conversion is handled in a predictable and transparent way. For a deeper dive, you can explore the various relationship types in the OMOPHub API documentation.
Validation, QA, and Performance Tuning
Getting an automated icd10 to icd9 conversion pipeline up and running is a huge step, but the job isn't done. The real challenge—and where the value truly lies—is making sure the output is both accurate and efficient enough to handle real-world data volumes.
Once your ETL is live, your focus has to shift. It's all about validation, quality assurance (QA), and performance tuning. Without this, you risk letting subtle mapping errors silently corrupt your entire dataset, which could lead to basing critical analytics on shaky ground.
A Practical Approach to Quality Assurance
A good QA process isn't about manually checking every single code. That’s just not feasible. Instead, it’s about implementing a smarter strategy to verify the integrity of the conversion at a high level and catch systemic problems early.
Here’s a checklist that works well in practice:
- Spot-Check High-Frequency Codes: Pull a list of your top 20 most common ICD-10 codes. Manually look up their ICD-9 mappings using a trusted source like the ATHENA vocabulary browser. This simple check ensures the data points you see most often are correct.
- Compare Cohort Sizes: Before and after the conversion, run counts on a few key patient cohorts, like those with Type 2 diabetes or hypertension. If you see a major, unexplained drop or spike in the cohort size, it’s a huge red flag that a mapping rule is likely off.
- Audit for Unmapped Codes: Your pipeline should be logging every ICD-10 code that doesn't map to an ICD-9 equivalent. Make a habit of reviewing this log. It’s the fastest way to spot gaps in the vocabulary or uncover quality issues in your source data.
The goal of QA isn't perfection on the first try. It's about building a transparent, iterative process. You need a system where you can confidently find, diagnose, and fix mapping issues as they pop up. That’s how you build long-term trust in the data.
Optimizing for Performance at Scale
When you're dealing with millions (or billions) of records, making a separate API call for every single code is a recipe for a slow, expensive pipeline. Performance tuning is non-negotiable.
One of the most powerful strategies is to introduce a local caching layer. The first time your script looks up an ICD-10 code, save the resulting ICD-9 mapping in a local key-value store like Redis or even just a simple dictionary. For every subsequent time that same code appears, you can grab the mapping from your lightning-fast local cache instead of making another network request.
Considering how often the same codes repeat in health data, this single technique can slash your API calls by over 90%.
Finally, always make sure your vocabulary mappings are current. Vocabularies evolve, and using an outdated version can compromise your accuracy. You can learn more about OHDSI’s methodology in their guide to vocabulary versioning and updates. This combination of rigorous QA and smart optimization is what makes a conversion process truly reliable and scalable.
Common Questions When Converting ICD Codes
Even with the best tools and a solid plan, you're bound to run into a few head-scratchers during an icd10 to icd9 conversion. Let's walk through some of the questions that come up most often for developers and data engineers working with OHDSI vocabularies and the OMOPHub API.
Is There Always a Perfect One-to-One Match?
Honestly, no. It's one of the first things you learn on a project like this. Expecting a direct, one-to-one match for every code is a recipe for frustration.
ICD-10 is far more granular than ICD-9, so it's common for a highly specific ICD-10 code to map back to a more general ICD-9 code. Sometimes, there’s just no direct equivalent at all. Your ETL logic needs to be built with this reality in mind. A solid strategy is to log any "no match" cases for later review instead of letting them halt the entire pipeline. This keeps the data flowing while giving you a clear list of records that might need a second look.
What’s the Deal with Standard vs. Non-Standard Concepts?
This is a core concept in the OHDSI world, and getting your head around it is crucial. Think of a standard concept as the "true north" for a clinical idea—the single, preferred way to represent it, usually a SNOMED CT code for something like "Type 2 diabetes mellitus."
Non-standard concepts are the source codes you start with, like your ICD-10 or ICD-9 codes. These codes 'Map to' a standard concept. So, when you're converting from ICD-10 to ICD-9, you're really following a path from one non-standard concept, through a standard concept, to another non-standard concept. Getting comfortable with this model is key to making sense of the vocabulary tables. For a deeper dive, the /concepts/search endpoint documentation on the OMOPHub docs site is a great resource.
The real power here is in the harmonization. By routing everything through a standard concept, you ensure that different source codes for the same clinical event all point to the same place. That’s a game-changer for accurate analytics.
How Do I Keep Up with Vocabulary Updates?
Medical vocabularies are always evolving. New codes are added, old ones are retired, and keeping up with these changes manually is a massive chore. You have to download the new files, load them into your database, and validate everything—it’s a huge operational headache.
This is where a managed service like the OMOPHub API really shines. It completely abstracts away all that backend maintenance. The platform handles the vocabulary updates for you, so your API calls are always hitting the latest official ATHENA release. Your team can focus on building the pipeline, not managing vocabulary files. You can check out the SDKs for this on the OMOPHub Python and OMOPHub R GitHub pages.
Tired of the infrastructure burden of managing vocabulary databases? With OMOPHub, you get instant REST API access to the full OHDSI ATHENA vocabularies. You can start building your conversion pipelines in minutes, not weeks. Ship faster and with more confidence by visiting https://omophub.com.


