The Ultimate Guide to ICD 9 DX Code Lookup with API Examples
An icd 9 dx code lookup is how developers can pull detailed information for a specific International Classification of Diseases, Ninth Revision, diagnosis code. It's a fundamental operation when you're working with historical healthcare data, turning a simple code string like '250.00' into a structured object packed with its official name, vocabulary, and unique identifiers.
Your Quick Start Guide to ICD 9 DX Code Lookup
Looking up an ICD-9 diagnosis code is usually the first thing you'll do in any data pipeline that touches legacy medical records. Before you can even think about complex mappings or crosswalks, you have to validate the code and get its basic details.
The OMOPHub API makes this straightforward. It lets you query for a specific concept and receive a standardized response, which is crucial for building dependable ETL jobs and analytics tools. If you've worked with other medical coding systems, this approach will feel a lot like doing an NDC code lookup.

Performing a Basic Lookup
The most direct route for an icd 9 dx code lookup is the /concept endpoint. This endpoint is built specifically for fetching the full details of a single concept when you already know its source code and vocabulary.
Here’s the typical workflow:
- Authentication: Your application will need to pass a valid API key with every request to authenticate with the OMOPHub service.
- API Call: You'll make a GET request to the
/conceptendpoint, passing theconcept_codeandvocabulary_idas query parameters. - Response Handling: The API will return a JSON object containing the detailed attributes of that concept, ready for your application to parse and use.
This simple, direct query is the building block for more advanced operations. For instance, you could easily write a script to loop through a list of ICD-9 codes from a source file, performing a lookup on each one to enrich your dataset.
Tip: Always specify the
vocabulary_idas'ICD9CM'in your request to avoid ambiguity and ensure you're querying the correct vocabulary. For more details on query parameters, see the OMOPHub API documentation. For SDK-specific examples, check out the OMOPHub repositories for Python and R.
Why ICD-9 Still Matters in a Post-Transition World
Even though the healthcare industry has moved on to ICD-10, the ghost in the machine is still ICD-9. For any developer or data engineer working with health data, an icd 9 dx code lookup isn't some obscure task—it's a frequent, practical necessity. If you touch any patient dataset originating before October 2015, you're almost guaranteed to encounter ICD-9-CM codes.
This legacy data is far from obsolete; it's a goldmine. It holds decades of clinical encounters, vital research information, and critical public health trends. To run a longitudinal study that bridges the transition date or to build a truly complete patient history, you have to be able to interpret this older information correctly. Simply dropping it isn't an option, as that would throw away priceless context and inject serious bias into your analysis.
The Scale of the Transition
The switch from ICD-9-CM to ICD-10-CM on October 1, 2015, was a seismic event in health data. The old system, with its rigid structure and dated medical terms, was hitting a wall and couldn't keep up with modern medicine.
To give you a sense of the scale, ICD-9-CM contained roughly 17,000 total codes. ICD-10 blew that up to over 155,000, enabling a much higher degree of detail and specificity. You can learn more about the scope of this massive healthcare data overhaul.
This huge jump in the number of codes is precisely why you often can't find a simple one-to-one match between the two systems. Performing an icd 9 dx code lookup is just the first step in a much more nuanced translation process that demands careful attention to preserve data integrity.
Navigating Data Continuity Challenges
For developers designing ETL pipelines, this historical split presents a significant hurdle. Your systems effectively need to be bilingual, fluent in both the old and new coding schemes. This isn't just about looking up codes; it's about understanding their relationships and mapping them accurately to a modern, unified vocabulary like SNOMED CT.
A robust data pipeline must treat ICD-9 not as an outdated relic but as a crucial component of historical data. The goal is to create a continuous, harmonized dataset that bridges the pre- and post-2015 eras without losing valuable clinical information along the way.
Grasping the modern-day relevance of ICD-9 becomes easier when you look at real-world applications. Many systems still rely on this historical data, as you'll see when exploring healthcare use cases for data processing. These examples drive home the persistent need for tools that can accurately handle and translate legacy codes for today's analytics platforms.
Mastering Advanced Search and Filtering
A simple icd 9 dx code lookup for a single term is one thing, but real-world data pipelines demand more muscle. You’ll often need to find concepts without knowing the exact code, weed out irrelevant results, or pinpoint codes based on very specific properties. This is precisely where the OMOPHub API’s advanced search parameters come into play.
Executing a more complex search is what lets you build dynamic, intelligent applications. Instead of hardcoding a list of known ICD-9-CM codes, your application can discover them on the fly using keywords like "diabetes" or other clinical criteria. This is incredibly useful for building interactive search UIs or for powering automated ETL jobs that have to classify messy, unpredictable lists of source codes.
Constructing Sophisticated Queries
The OMOPHub API enhances the /concepts endpoint with several query parameters you can chain together to fine-tune your search. This analytical approach gives you the power to find the exact concepts you need with surgical precision, which means a lot less post-processing work on your end.
Here are a few of the most common filtering parameters:
query: This performs a keyword search across concept names and their synonyms. A search likequery=diabeteswill pull back all concepts related to the term.concept_class_id: Use this to restrict results to a specific class, like '4-dig billing code' or '5-dig billing code'. It's a great way to filter by code granularity.standard_concept: This lets you filter for standard ('S'), non-standard ('N'), or classification ('C') concepts. This is absolutely critical when you need to isolate source codes that require mapping.
These parameters transform a basic lookup into a flexible discovery tool. For a complete rundown of every available parameter, check out the official OMOPHub documentation.
Tip: When building a search feature, combine the
queryparameter with filters likevocabulary_id='ICD9CM'andstandard_concept='N'. This setup ensures your application specifically searches for relevant non-standard ICD-9-CM source codes—the perfect candidates for mapping to modern OMOP standards.
To get your hands on the SDKs and start coding, dive into the examples available in the official GitHub repositories for both Python and R. Those repos are packed with practical snippets for building out your first advanced icd 9 dx code lookup function.
Mapping ICD 9 Codes to Modern OMOP Standards
An icd 9 dx code lookup is just the first step. The real analytical value comes from what you do with that code next—transforming it. In the world of healthcare analytics, source codes like ICD-9-CM are essentially different dialects. To run any kind of large-scale analysis, you have to translate them into a common language. That's precisely what the OMOP Common Data Model does by mapping source codes to a unified set of standard concepts, usually from SNOMED CT.
This mapping isn't just a technical detail; it's the entire foundation of modern health data interoperability. It's what allows a researcher to ask for "Type 2 diabetes" and pull records from a dozen different sources, even if one used ICD-9, another used ICD-10, and a third used some proprietary system. Without this standardization, building a cohesive patient cohort would be a nightmare of manual, error-prone work. If you're new to this framework, you can learn more about the OMOP data model and its components.
Traversing Concept Relationships
The magic behind this translation lies in the concept relationships. Inside the OMOP vocabulary tables, you'll find explicit links that show how a non-standard source code (like our ICD-9-CM code) connects to its standard equivalent. The most important relationship you'll work with is 'Maps to'.
With the OMOPHub API, you can follow these connections programmatically. Instead of just looking up a code, you're querying for a source concept and then asking for its relationships to discover the standard concept on the other side. This is how you build robust, automated ETL pipelines that can turn messy legacy data into a clean, analysis-ready format.
Here's a high-level look at how it works:
- Lookup the Source Concept: First, you run an
icd 9 dx code lookupfor your original ICD-9-CM code (e.g., '250.00'). - Query Relationships: With the source concept's ID in hand, you hit the
/relationshipsendpoint to pull all its connections. - Filter for Standard Mapping: From that list, you just need to find the one where the
relationship_idis 'Maps to' and the target concept hasstandard_concept = 'S'.
Tip: The concept you get back is the standardized equivalent you should populate your OMOP CDM tables with. Sticking to this process ensures your data will play nicely with other OMOP-formatted datasets across the globe.
Handling Complex Mappings
It's not always a simple one-to-one journey. While many ICD-9 codes map cleanly, you'll inevitably run into more complicated scenarios. A single ICD-9 code might map to multiple standard concepts (a one-to-many relationship), or worse, it might not map to anything at all (one-to-zero).
Your application's logic has to be ready to handle this ambiguity. For one-to-many mappings, you might need to use other clinical details from the source record—like lab values or notes—to decide which standard concept is the best fit. When a code doesn't map at all, your pipeline could flag the record for a human to review or fall back to mapping it to a broader, less specific parent concept. You can find some excellent code examples for navigating these edge cases in the official OMOPHub Python SDK and R SDK repositories.
Navigating the ICD-9 to ICD-10 Crosswalk with Precision
For anyone working with longitudinal health data, translating legacy ICD-9-CM codes into their modern ICD-10-CM counterparts is a fundamental task. While an icd 9 dx code lookup is the starting point, the real goal is to bridge the data gap created by the 2015 transition. For developers, knowing how to access this crosswalk programmatically is non-negotiable.
An API gives you the power to find all potential ICD-10-CM mappings for a given ICD-9-CM code. This is absolutely critical for any project touching data from before and after the switchover, whether you're migrating databases, updating clinical decision support rules, or conducting research that spans multiple years.
The shift from ICD-9 to ICD-10 was more than just a code update; it fundamentally changed the granularity of health data analysis. The specificity baked into ICD-10 codes—capturing details like laterality and severity—opened the door for much richer insights. At the same time, this created significant data continuity headaches. Public health organizations had to completely redefine case definitions and build consensus on how to map historical ICD-9 data into the new system, which really underscores the need for a reliable crosswalk. You can get a much deeper look into these issues in research covering the ICD-9 to ICD-10 evolution.
Interpreting Crosswalk Results
When you look up an ICD-10 equivalent for an ICD-9 code, you'll quickly find it’s not always a straightforward, one-to-one swap. Your code needs to be ready to handle a few different mapping scenarios.
- One-to-One Mapping: This is the clean, ideal case where one ICD-9 code maps directly to a single ICD-10 code.
- One-to-Many Mapping: It's very common for a single, less specific ICD-9 code to branch out into several more granular ICD-10 codes.
- Combination Mapping: In some complex cases, you might need multiple ICD-9 codes to accurately capture the full meaning of a single ICD-10 concept.
This diagram gives a high-level view of the process, showing how an older ICD-9 code can be mapped to a standardized vocabulary through an API.
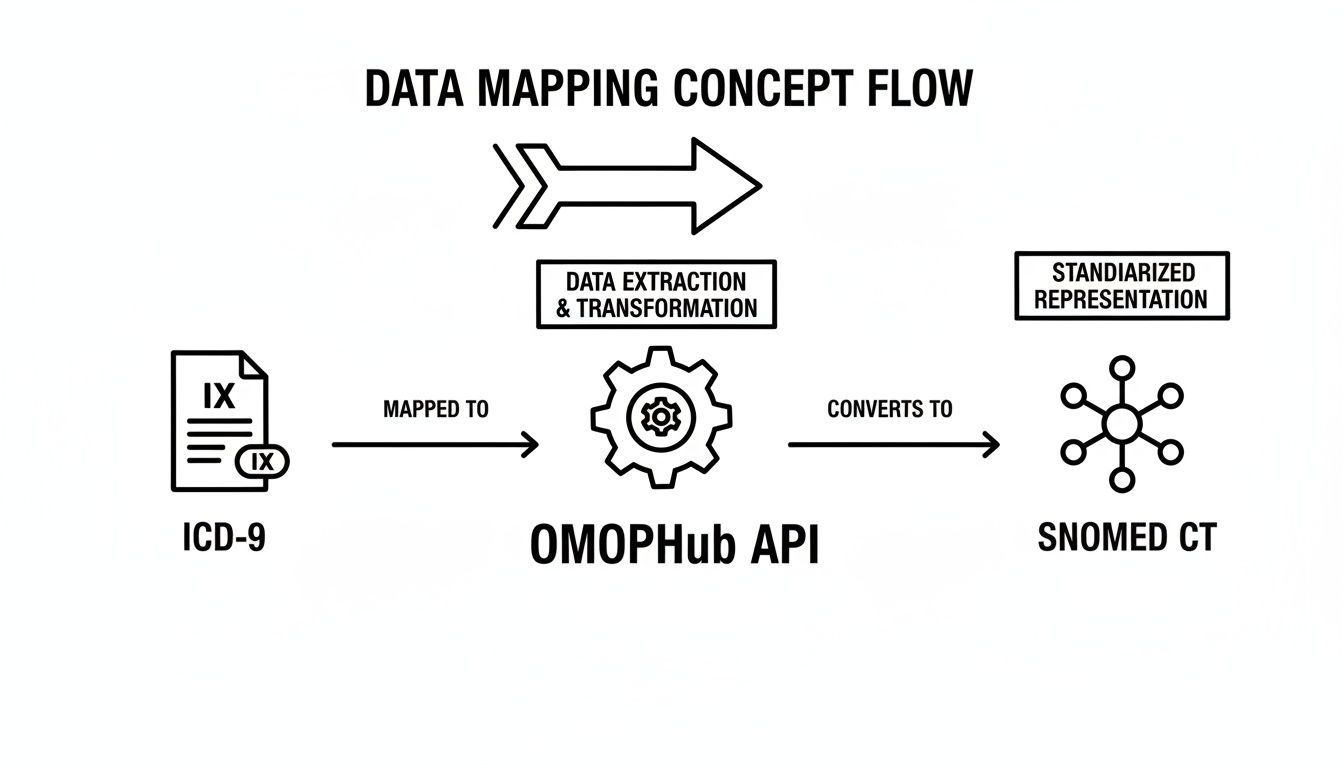
As the diagram shows, the OMOPHub API can serve as the intermediary that translates these legacy codes into a modern, interoperable format like SNOMED CT. While we're focused on the ICD-9 to ICD-10 crosswalk here, the same core principles apply when you need to go in the other direction. For more on that, check out our guide on ICD-10 to ICD-9 conversion.
Best Practices for Data Versioning and Provenance
When you're running an icd 9 dx code lookup, getting the right code is only half the battle. You also need to guarantee data integrity through solid versioning and provenance. Healthcare vocabularies aren't static; codes get added, retired, or redefined all the time. If your application needs auditable and reproducible results, knowing which version of a vocabulary was used is non-negotiable.
OMOPHub handles this by versioning the entire vocabulary set with each official ATHENA release. This simple but powerful approach ties every API call you make to a specific, unchangeable version of the data—a critical feature for building compliant, enterprise-grade systems.
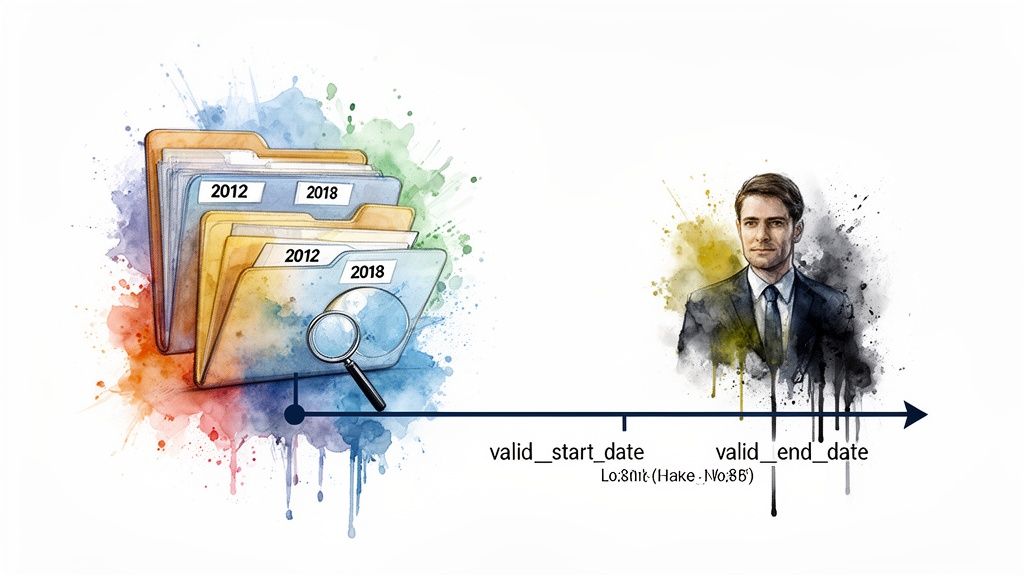
Aligning Data with Vocabulary Versions
If you're dealing with historical datasets, you have to align your data with the vocabulary version that was active when the data was created. It’s a common pitfall. A code that was perfectly valid in 2010 might be deprecated in a 2020 release. Using the wrong version is a recipe for incorrect mappings and flawed analytics.
The OMOPHub API gives you granular control by letting you specify a vocabulary version right in your request. This gives you command over your data's temporal context, which is especially important for longitudinal studies that span multiple years.
Ensuring Historical Accuracy with Date Ranges
A cornerstone of good data provenance is knowing a concept's period of validity. The OMOP vocabulary makes this straightforward with two essential fields: valid_start_date and valid_end_date. Together, they define the exact timeframe during which a concept is considered active and standard.
Here are a couple of practical tips for working with these fields:
- Check the
valid_end_date: When looking up a code, always verify that thevalid_end_dateis either far in the future (usually '2099-12-31' for currently active codes) or falls after the timestamp of your source data. - Filter out invalid concepts: Make it a standard practice to filter out any concepts where the
valid_end_datehas already passed. These are retired codes and shouldn't be used for any new mappings.
Tip: By consistently checking these date fields, you build a more robust and auditable data pipeline. This habit ensures your
icd 9 dx code lookupoperations are not just accurate at a single point in time but are historically correct—the foundation of any reproducible research.
For a deeper dive into versioning, check out the official OMOPHub documentation or explore the SDKs for Python and R.
Frequently Asked Questions About ICD-9 Lookups
Working with an icd 9 dx code lookup can throw a few curveballs, even for experienced developers. This section gets right to the point, tackling some of the most common questions we see. Think of it as a quick troubleshooting guide for your data handling workflows.
Why Do I Still Need to Look Up ICD-9 Codes?
It’s a fair question. After all, the US healthcare system officially moved to ICD-10 back in 2015. The reality is that a tremendous amount of valuable historical data—everything from old electronic health records (EHRs) and claims databases to longitudinal clinical studies—is still coded in ICD-9-CM.
If your project involves any data recorded before October 2015, you absolutely need a reliable way to look up and make sense of these legacy codes. It's the only way to ensure data continuity. Whether you're building a patient's complete health history or training a machine learning model on decades of data, mapping these older codes to modern standards like SNOMED CT is essential for maintaining analytical integrity.
How Do I Handle an ICD-9 Code That Maps to Multiple ICD-10 Codes?
You'll run into this all the time. ICD-10 introduced a much higher level of specificity, so it's common for a single, more general ICD-9 code to branch out into several more precise ICD-10 codes. Your application logic needs a clear plan for this.
Here’s how most teams approach it:
- Look for More Context: The ideal solution is to pull in other clinical details from the source record. Physician notes, lab results, or laterality data (e.g., "left knee" vs. "right knee") can often point you to the single correct ICD-10 code.
- Flag for a Human Review: If there's just not enough context in the data to make an automated decision, your ETL process should flag the record. A clinical expert can then review it manually.
- Map to a Broader Concept: As a last resort, you can map the code to a less specific "parent" concept in the hierarchy that accurately covers all the potential child codes.
What’s the Difference Between a Source and a Standard Concept in OMOP?
Understanding this distinction is fundamental to working with the OMOP Common Data Model. The concept is pretty straightforward once you get it.
A source concept is simply the code as it appears in the original data. For instance, if the raw data has the ICD-9-CM code '250.00' for Type 2 diabetes, that's your source concept.
A standard concept, on the other hand, is the preferred, standardized code that the OMOP CDM uses for analysis, which usually comes from a robust reference vocabulary like SNOMED CT. The whole point of the model is to map all those different source codes (from ICD-9, ICD-10, etc.) to a single set of standard concepts. This is what makes it possible to run consistent, large-scale analytics across diverse datasets. Of course, when handling any health data, it's not just about the technicals; knowing the risks of HIPAA non-compliance is critical.
Accelerate your healthcare data projects with developer-first tools. With OMOPHub, you can eliminate the overhead of managing vocabulary databases and get instant, compliant API access to SNOMED, ICD, and more. Get started today at https://omophub.com.


