Top 6 Ways: inclusion criteria example for Clinical Cohorts in 2026

Defining a study cohort is the bedrock of reproducible clinical research. While protocols outline the 'who,' the real challenge lies in translating those rules into precise, executable logic against real-world data like the OMOP Common Data Model. A single, poorly defined inclusion criteria example can invalidate results, introduce bias, and waste months of effort. This precision is critical; just as success in hiring requires proactively building a crystal-clear profile of your ideal candidate, robust research demands an equally meticulous definition of its participants.
This article moves beyond theory to provide concrete examples that bridge the gap between clinical protocol and code. We will break down 8 common inclusion criteria, detailing their strategic rationale, common implementation pitfalls, and practical execution snippets using the OMOPHub SDKs for Python and R. Readers will learn replicable strategies for defining everything from simple age ranges to complex medication exposure windows. Our goal is to equip data scientists and clinical researchers with the tactical insights needed to build valid, scalable, and clinically meaningful cohorts, accelerating their analytics pipelines and ensuring the integrity of their findings.
1. Age Range Specifications
Age range specifications are a foundational inclusion criteria example, defining the minimum and maximum age for participant eligibility. This criterion is crucial for ensuring that a study cohort is demographically appropriate for the research question, whether it's a COVID-19 vaccine trial for adults (18+ years), a pediatric medication safety study (0-18 years), or a geriatric fall prevention analysis (65+ years).
In observational research using the OMOP Common Data Model, age is not a static field but is dynamically calculated. It's typically derived from the birth_datetime in the PERSON table relative to a specific event, such as a condition diagnosis, drug exposure start date, or a defined study index date.
Strategic Analysis
Implementing age criteria requires careful consideration of the temporal context. The key decision is defining the "as of" date for the age calculation. Is it age at the first encounter, age at the cohort index date, or age at the end of the observation period? This choice directly impacts cohort composition and must be clearly documented in the study protocol.
For example, a study on early-onset Type 2 Diabetes might define its cohort as "persons aged 18-40 at the time of their first T2D diagnosis." An incorrect calculation, like using the person's current age in the database, would erroneously include patients diagnosed in their 30s who are now in their 50s.
Key Insight: Precise age calculation is not just about filtering; it's about establishing a clinically relevant temporal anchor for the entire cohort. The logic must account for the relationship between the participant's birth date and the specific clinical event defining their entry into the study.
Actionable Takeaways & Implementation
To reliably implement this inclusion criteria example, data teams must standardize their age calculation logic and perform rigorous data quality checks.
- Tip 1: Standardize Calculation Logic: Use the
person.birth_datetimefield and calculate age relative to a defined index date from another table (e.g.,condition_occurrence.condition_start_date). Document this logic clearly. - Tip 2: Handle Boundary Cases: Test your cohort definition logic for edge cases, such as individuals who turn the minimum required age on the day of the index event.
- Tip 3: Implement Data Quality Checks: Before applying the age filter, run checks for missing
birth_datetimevalues and screen for implausible ages (e.g., >120 years or negative ages), which can indicate data entry errors.
Here is a simplified pseudocode snippet for identifying a pediatric cohort (age < 18) at the time of their first qualifying condition:
Pseudocode for OMOP-based age filtering
Assumes 'person_df' and 'condition_df' are dataframes
Join PERSON and CONDITION_OCCURRENCE tables
merged_df = pd.merge(person_df, condition_df, on='person_id')
Calculate age at condition start date
merged_df['age_at_condition'] = ( merged_df['condition_start_date'].dt.year - merged_df['birth_datetime'].dt.year )
Filter for the pediatric cohort
pediatric_cohort = merged_df[merged_df['age_at_condition'] < 18]
2. Diagnosis Code Requirements (ICD-10-CM, SNOMED CT)
Diagnosis code requirements are a fundamental inclusion criteria example that uses standardized medical vocabularies like ICD-10-CM and SNOMED CT to identify patients with specific conditions. This criterion is vital for creating clinically coherent cohorts, from broad studies on heart failure (ICD-10-CM I50.*) to highly specific research on Type 2 diabetes with renal complications (a combination of SNOMED CT concepts).
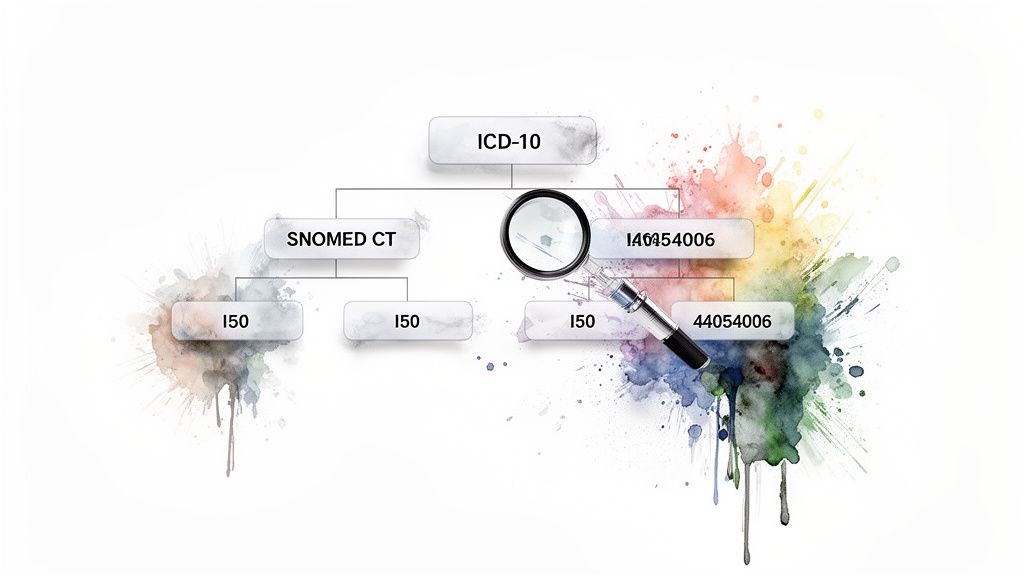
In the OMOP Common Data Model, these diagnoses are stored in the CONDITION_OCCURRENCE table, with the condition_concept_id linking to the standard vocabulary concept in the CONCEPT table. This standardized structure allows researchers to build precise, reproducible definitions for conditions of interest.
Strategic Analysis
The strategic challenge lies in creating a "concept set" that accurately captures the clinical intent of the study. A simple code search is often insufficient. For instance, a cohort for "Type 2 diabetes" must include not only the primary SNOMED CT code (44054006) but also its numerous descendants representing more specific forms of the disease. This requires traversing the vocabulary's hierarchical relationships.
Furthermore, deciding on the vocabulary and hierarchy depth is a critical protocol decision. Should a heart failure study only include I50 codes, or should it expand to related concepts mapped from other vocabularies? The choice impacts both the sensitivity and specificity of the cohort definition. Using a tool like the OMOPHub API simplifies this by programmatically identifying and including all relevant descendant concepts, removing manual effort and potential errors.
Key Insight: Building a robust diagnosis-based cohort is less about finding a single code and more about defining a comprehensive and logically consistent set of concepts. The strategy must account for vocabulary hierarchies and cross-vocabulary mappings to ensure the cohort is clinically sound and reproducible.
Actionable Takeaways & Implementation
To effectively implement this inclusion criteria example, researchers and data engineers must leverage vocabulary tools and establish clear documentation practices.
- Tip 1: Use Hierarchical Expansion: Instead of manually listing codes, use programmatic tools to find a standard concept and include all its descendants. The OMOPHub Python SDK's
concepts.get_descendants()function automates this crucial step. - Tip 2: Document Your Concept Set: Clearly list all included
concept_ids, the vocabulary version (e.g., ATHENA release date), and the rationale for the hierarchy depth in your study protocol to ensure reproducibility. - Tip 3: Validate Mappings: When using codes from multiple vocabularies (e.g., ICD-10-CM and SNOMED CT), verify that the mappings align with your clinical intent. A source ICD code may map to a broader or narrower standard SNOMED concept.
Here is a simplified snippet using the OMOPHub Python SDK for identifying a Type 2 diabetes cohort using a base SNOMED CT concept and its descendants:
# Code example using the OMOPHub Python SDK
# See SDK documentation: https://docs.omophub.com/sdk/python
from omophub.sdk import OMOPHub
# Initialize the client
client = OMOPHub()
# Define the base concept for Type 2 Diabetes (SNOMED CT)
t2dm_concept_id = 44054006
# Get all descendant concept IDs
descendants = client.concepts.get_descendants(concept_id=t2dm_concept_id)
t2dm_concept_set_ids = [c.concept_id for c in descendants]
t2dm_concept_set_ids.append(t2dm_concept_id) # Include the base concept itself
# Now use this t2dm_concept_set_ids to filter the CONDITION_OCCURRENCE table
# e.g., t2dm_cohort = condition_df[condition_df['condition_concept_id'].isin(t2dm_concept_set_ids)]
3. Laboratory Test Value Thresholds
Laboratory test value thresholds are a quantitative inclusion criteria example that defines participant eligibility based on specific biomarker measurements. This is essential for creating clinically homogenous cohorts, such as requiring an HbA1c level of ≥6.5% for a Type 2 Diabetes study (LOINC 4548-4) or an eGFR <60 mL/min/1.73m² for a Chronic Kidney Disease analysis (LOINC 33914-3).
In the OMOP Common Data Model, these values are stored in the MEASUREMENT table. The criteria involve linking a specific test, identified by a measurement_concept_id (often mapped from a LOINC code), to its numeric result (value_as_number) and corresponding units (unit_concept_id).
Strategic Analysis
The primary challenge with lab values is variability in units and timing. A study might need the first qualifying HbA1c value post-index, the most recent LDL-C value, or any eGFR value below a threshold within a one-year window. The logic must precisely define this temporal relationship and handle unit conversions (e.g., mg/dL vs. mmol/L for glucose).
For instance, defining a cohort with "hyperlipidemia controlled by statins" could require identifying persons with an LDL-C result <100 mg/dL after their first statin prescription. Simply finding any low LDL-C value in their history would incorrectly include individuals who had low cholesterol before ever needing treatment, fundamentally misrepresenting the study population.
Key Insight: Implementing laboratory-based inclusion criteria requires a three-part logic: identifying the correct test via its concept ID, applying the numeric threshold, and anchoring the measurement event to a specific clinical timeline (e.g., baseline, post-treatment, most recent).
Actionable Takeaways & Implementation
Reliable implementation of this inclusion criteria example hinges on robust concept mapping, unit standardization, and clear temporal logic. The OMOPHub SDKs can streamline the process of finding concepts for tests like HbA1c.
- Tip 1: Standardize Units: Before applying filters, create logic to convert different units for the same lab test to a single standard (e.g., convert all glucose measurements to mg/dL). Document this conversion protocol clearly.
- Tip 2: Define Temporal Logic: Explicitly state whether the criterion applies to the baseline value, the first value, the last value, or any value within a specific observation window relative to the cohort index date.
- Tip 3: Perform Sanity Checks: Implement data quality checks to flag or exclude physiologically implausible values (e.g., HbA1c >25%, eGFR >200) that likely represent data entry errors before applying the inclusion threshold.
This simplified Python code shows how to identify a cohort with an HbA1c value indicating diabetes, using the OMOPHub Python SDK to find the concept ID.
# Code example using the OMOPHub Python SDK
from omophub.sdk import OMOPHub
# Initialize the client and assume 'measurement_df' is a Pandas DataFrame
client = OMOPHub()
# Use OMOPHub SDK to find the concept ID for HbA1c
hba1c_concepts = client.concepts.search(query="HbA1c", vocabulary_id=["LOINC"])
hba1c_concept_id = hba1c_concepts[0].concept_id # Example: 3004410 for 'Hemoglobin A1c/Hemoglobin.total'
# Filter for the specific lab test
hba1c_measurements = measurement_df[
measurement_df['measurement_concept_id'] == hba1c_concept_id
]
# Apply the value threshold for the inclusion criteria
# Note: Unit handling (e.g., ensuring values are in %) is critical here.
diabetes_cohort = hba1c_measurements[
hba1c_measurements['value_as_number'] >= 6.5
]
4. Medication Exposure Windows (RxNorm)
Medication exposure is a powerful inclusion criteria example that defines eligibility based on whether a participant has been prescribed or dispensed specific drugs within a defined time frame. This criterion is essential for studies focusing on drug efficacy, safety, or adherence, such as identifying patients on a statin for a cardiovascular outcomes trial or those receiving a specific antibiotic class for a stewardship program.

In the OMOP Common Data Model, medication exposures are recorded in the DRUG_EXPOSURE table. The RxNorm vocabulary standardizes these records, mapping various drug codes (like NDCs) to a consistent hierarchical structure of ingredients, brand names, and dose forms. This allows researchers to reliably query for entire drug classes (e.g., all ACE inhibitors) or specific ingredients.
Strategic Analysis
The core challenge in implementing medication exposure criteria lies in defining the temporal window and the required level of drug specificity. Is a single prescription sufficient, or must exposure be continuous? Are we interested in a specific brand name or any ingredient in a therapeutic class? The answers to these questions dictate the complexity of the cohort logic.
For instance, a study on ACE inhibitor safety might define its cohort as "persons with at least one dispensing event for any drug descending from RxNorm ingredient 39980 (Lisinopril) within 90 days prior to the index date." A more stringent study might require "continuous use," defined as having no gap greater than 30 days between the end of one prescription's days supply and the start of the next.
Key Insight: Effective medication exposure criteria are not just about finding a drug code; they are about defining a clinically meaningful relationship between the drug, the patient, and the study's timeline. This requires precise temporal logic and a deep understanding of the RxNorm hierarchy.
Actionable Takeaways & Implementation
To successfully apply this inclusion criteria example, teams need to leverage vocabulary tools for concept set creation and meticulously define their temporal logic.
- Tip 1: Build Robust Concept Sets: Use a tool like the OMOPHub API to explore RxNorm ingredient hierarchies and programmatically build concept sets for entire drug classes. See the OMOPHub Python SDK for examples.
- Tip 2: Define Explicit Exposure Windows: Clearly document the temporal logic, such as "any prescription fill within 180 days prior to index" versus "at least two fills for the same ingredient in the year before index."
- Tip 3: Account for Medication Gaps: When assessing continuous use, define a permissible gap (e.g., 30, 60, or 90 days) between prescriptions to account for real-world adherence patterns and avoid prematurely excluding eligible patients.
Here is a simplified pseudocode snippet for identifying a cohort with recent statin exposure using an RxNorm concept set:
Pseudocode for OMOP-based drug exposure filtering
Assumes 'drug_exposure_df' is a dataframe and 'statin_concept_ids' is a list of RxNorm IDs
Define index date and lookback window
index_date = '2023-01-01' lookback_days = 90 start_window = index_date - timedelta(days=lookback_days)
Filter for drug exposures within the concept set
statin_exposures = drug_exposure_df[ drug_exposure_df['drug_concept_id'].isin(statin_concept_ids) ]
Filter for exposures within the lookback window
recent_statin_cohort = statin_exposures[ (statin_exposures['drug_exposure_start_date'] >= start_window) & (statin_exposures['drug_exposure_start_date'] <= index_date) ]
5. Observation Period and Data Availability
Observation period and data availability serve as a critical inclusion criteria example to ensure patients have sufficient longitudinal data for meaningful analysis. This criterion validates that individuals have an adequate history for baseline characteristic assessment and enough follow-up time to capture study outcomes. It's fundamental for studies requiring continuous enrollment, such as comparing long-term treatment effects or validating predictive models.
In the OMOP Common Data Model, the OBSERVATION_PERIOD table directly supports this. Each record represents a span of time during which a person has continuous observation in the source data, defined by observation_period_start_date and observation_period_end_date. This allows researchers to reliably select cohorts with specific windows of data visibility.
Strategic Analysis
Implementing observation period criteria requires defining the data window relative to a specific anchor point, usually the cohort index date. The key decision is how much pre-index (baseline) and post-index (follow-up) observation is necessary to answer the research question. Insufficient baseline data can lead to unmeasured confounding, while inadequate follow-up can result in missed outcomes.
For example, a study on the long-term side effects of a new medication might require "at least 365 days of continuous observation prior to the first drug exposure and at least 730 days of continuous observation after." This ensures a robust baseline profile can be built and that there is a sufficient time window to observe potential adverse events.
Key Insight: Observation period criteria are not just about data quantity; they are about guaranteeing the temporal integrity required to establish causality and measure longitudinal outcomes accurately. The definition of "continuous" is crucial and must account for permissible gaps in data capture.
Actionable Takeaways & Implementation
To effectively apply this inclusion criteria example, data teams must standardize how they define and verify observation windows relative to cohort index events.
- Tip 1: Anchor to an Index Date: Always define observation windows relative to a specific event (e.g.,
condition_start_date,drug_exposure_start_date). Use theOBSERVATION_PERIODtable to confirm the index date falls within a valid period. - Tip 2: Define and Document Gap Logic: Clearly specify what constitutes a permissible gap in observation (e.g., 30 or 60 days). This logic is vital for accurately constructing continuous observation periods from source data.
- Tip 3: Validate with Sensitivity Analyses: Test the robustness of your findings by varying the observation period requirements. For example, compare the results of a cohort with ≥1 year of follow-up versus one with ≥2 years to understand the impact on the outcome.
Here is a simplified pseudocode snippet for selecting a cohort with at least 365 days of prior observation:
Pseudocode for OMOP-based observation period filtering
Assumes 'cohort_df' has person_id and index_date
Assumes 'obs_period_df' has person_id, start_date, end_date
Join cohort with their observation periods
merged_df = pd.merge(cohort_df, obs_period_df, on='person_id')
Ensure the index date falls within an observation period
merged_df = merged_df[ (merged_df['index_date'] >= merged_df['start_date']) & (merged_df['index_date'] <= merged_df['end_date']) ]
Calculate days of prior observation
merged_df['prior_obs_days'] = ( merged_df['index_date'] - merged_df['start_date'] ).dt.days
Filter for cohort with at least 1 year of prior observation
final_cohort = merged_df[merged_df['prior_obs_days'] >= 365]
6. Procedure Code Inclusion (CPT, SNOMED CT)
Procedure code inclusion criteria use standardized codes to identify patients who have undergone specific surgical, diagnostic, or therapeutic interventions. This inclusion criteria example is essential for studies focused on surgical outcomes, medical device efficacy, or intervention effectiveness. Vocabularies like CPT (Current Procedural Terminology) and SNOMED CT provide the necessary granularity to define cohorts, such as patients with a total knee arthroplasty (CPT 27447) or an aortic valve replacement (SNOMED 10190003).
In the OMOP Common Data Model, these events are stored in the PROCEDURE_OCCURRENCE table. Researchers build concept sets of relevant procedure codes to define their cohort, ensuring that participants have the qualifying clinical history required by the study protocol. This method allows for the creation of highly specific, clinically relevant patient groups.
Strategic Analysis
The primary challenge in using procedure codes is ensuring both completeness and precision. A study on knee replacement, for instance, must include not only the primary CPT code but also variants for revisions, partial replacements, or different surgical approaches. Conversely, a code for a general "joint procedure" would be too broad and introduce noise.
The strategic decision lies in defining the temporal relationship between the procedure and the study's index date. Is the procedure the index event itself? Or must it have occurred within a specific timeframe (e.g., "coronary artery bypass graft within the 30 days prior to cohort entry")? This temporal logic is critical for establishing a clear cause-and-effect timeline.
Key Insight: Building a robust procedure-based cohort is less about finding a single code and more about curating a comprehensive concept set that accounts for clinical variations while defining strict temporal rules relative to the study timeline.
Actionable Takeaways & Implementation
To effectively implement procedure-based criteria, teams must leverage vocabulary tools and establish clear rules for timing and hierarchy.
- Tip 1: Build Hierarchical Concept Sets: Use vocabulary tools like the OMOPHub SDK to build concept sets that include not just a specific code but also its descendants in the SNOMED hierarchy. This captures related procedures and prevents missed cases.
- Tip 2: Define Strict Temporal Windows: Clearly specify the timing of the procedure relative to the cohort index date (e.g., "procedure occurs on index date" or "procedure must occur in the 180 days before index").
- Tip 3: Validate with Clinical Context: After defining a cohort, review a sample of patient records or associated clinical notes to confirm the procedure code accurately reflects the intended clinical event. This helps catch coding inaccuracies or ambiguity.
The pseudocode below shows a simplified approach to identifying a cohort of patients who have undergone a total knee arthroplasty.
Pseudocode for OMOP-based procedure filtering
Assumes 'procedure_df' is a dataframe of PROCEDURE_OCCURRENCE
Define the concept set for Total Knee Arthroplasty (TKA)
In practice, this set would be larger, including related codes.
See OMOPHub docs for building concept sets: https://docs.omophub.com
tka_concept_ids = {2001325, 40488924} # Example concept IDs for TKA
Filter for procedures within the defined concept set
tka_cohort = procedure_df[ procedure_df['procedure_concept_id'].isin(tka_concept_ids) ]
7. Visit Type and Healthcare Setting Specifications
Visit type and healthcare setting specifications are an essential inclusion criteria example used to define the clinical context of patient encounters. This criterion ensures that the cohort is built from data generated in appropriate settings, such as including only inpatient hospitalizations for a hospital-acquired infection study (OMOP visit_concept_id = 9201) or restricting to outpatient office visits for a primary care medication adherence analysis.
In the OMOP Common Data Model, the VISIT_OCCURRENCE table captures these encounters. The visit_concept_id field is key, classifying the setting (e.g., Inpatient Visit, Emergency Room Visit, Outpatient Visit). This allows researchers to precisely target interactions relevant to their study question, whether it's acute care events or routine follow-ups.
Strategic Analysis
The strategic implementation of visit type criteria hinges on correctly mapping source system encounter types to standard OMOP visit concepts. A study’s validity can be compromised if, for example, an "observation stay" is misclassified as a standard "outpatient visit," as this would exclude patients who received significant, albeit brief, hospital-level care.
Consider a study on the efficacy of telehealth services. The inclusion criteria might be "persons with a video or telephone encounter post-2020." Simply filtering for any outpatient visit would be too broad. The cohort definition must specifically target concept IDs representing virtual care to ensure the analysis is based on the correct intervention setting.
Key Insight: Specifying the visit type is not just about filtering data; it is about defining the clinical and operational environment where the data was generated, which directly impacts the generalizability and interpretation of study findings.
Actionable Takeaways & Implementation
To reliably implement this inclusion criteria example, teams must standardize their visit mapping logic and validate the clinical context of the resulting cohort.
- Tip 1: Standardize Visit Mappings: Use resources like the OMOPHub documentation to understand and correctly map source encounter codes to standard
visit_concept_idandvisit_type_concept_idvalues. - Tip 2: Document Criteria Explicitly: Clearly state which visit types are included. For instance, specify if a cohort includes "inpatient plus observation stays" versus just "inpatient" to avoid ambiguity.
- Tip 3: Validate Visit Logic: Perform quality checks to confirm visit start and end dates are logical and fall within a patient's observation period. Check for plausible visit transitions, like an Emergency Room visit immediately preceding an Inpatient Stay on the same day.
Here is a simplified pseudocode snippet for identifying a cohort of patients with at least one inpatient hospitalization:
Pseudocode for OMOP-based visit filtering
Assumes 'visit_occurrence_df' is a dataframe
Standard OMOP Concept ID for Inpatient Visit is 9201
INPATIENT_CONCEPT_ID = 9201
Filter for all inpatient visits
inpatient_visits = visit_occurrence_df[ visit_occurrence_df['visit_concept_id'] == INPATIENT_CONCEPT_ID ]
Identify unique persons who have had an inpatient visit
inpatient_cohort_person_ids = inpatient_visits['person_id'].unique()
8. Exclusion Criteria Definition (Contraindications, Comorbidities)
Defining what makes a patient ineligible is a critical, albeit inverse, part of crafting inclusion criteria. This inclusion criteria example focuses on specifying contraindications or comorbidities that disqualify participants. It is essential for patient safety and study validity, ensuring individuals are not exposed to undue risk and that the cohort is appropriate for the research question, such as excluding pregnant individuals from certain drug trials or those with severe renal disease from studies of medications cleared by the kidneys.
In the OMOP Common Data Model, these exclusions are implemented by identifying records in tables like CONDITION_OCCURRENCE, DRUG_EXPOSURE, or MEASUREMENT that correspond to the specified contraindications. Researchers create "negative cohorts" based on concept sets representing these conditions and filter them out from the potential study population.
Strategic Analysis
The strategic challenge in defining exclusions lies in balancing scientific rigor with practical generalizability. Overly restrictive exclusions can create a "perfect" but highly niche cohort that doesn't reflect the real-world patient population, limiting the external validity of the findings. The key is to define precise clinical and temporal parameters for each exclusion.
For instance, excluding patients with "a history of cancer" is vague. A more robust definition would specify "any malignancy diagnosis, excluding non-melanoma skin cancer, within five years prior to the index date." This requires not just identifying the right diagnosis codes but also applying a specific lookback window relative to the patient's cohort entry date.
Key Insight: Effective exclusion criteria are not just a list of conditions to avoid; they are a precisely defined set of rules with clear temporal boundaries that protect participants and reduce confounding variables without unnecessarily sacrificing the study's generalizability.
Actionable Takeaways & Implementation
To robustly define and apply exclusions, data teams must build reusable concept sets and clearly document the logic, particularly the temporal constraints.
- Tip 1: Build Reusable Concept Sets: Use vocabulary tools to create and maintain standardized concept sets for common exclusions like severe renal disease or pregnancy. This ensures consistency across studies. For programmatic creation, see the OMOPHub SDK documentation.
- Tip 2: Define Clear Temporal Windows: Explicitly state the time window for each exclusion (e.g., "any diagnosis of heart failure in the 365 days prior to index"). This prevents ambiguity in cohort definition.
- Tip 3: Conduct Sensitivity Analyses: Assess the impact of your exclusion criteria by running the analysis on a cohort with relaxed exclusions. This can help quantify how specific criteria affect the final results and cohort size.
Here is a simplified pseudocode snippet for excluding patients with a record of severe renal disease prior to a study index date:
Pseudocode for OMOP-based exclusion filtering
Assumes 'potential_cohort_df' and 'condition_df' are dataframes
'severe_renal_disease_concept_ids' is a predefined set of codes
Isolate severe renal disease diagnoses
renal_diagnoses = condition_df[ condition_df['condition_concept_id'].isin(severe_renal_disease_concept_ids) ]
Identify patients with this diagnosis
patients_to_exclude = set(renal_diagnoses['person_id'])
Filter the main cohort to exclude these patients
final_cohort = potential_cohort_df[ ~potential_cohort_df['person_id'].isin(patients_to_exclude) ]
8-Point Inclusion Criteria Comparison
| Criterion | Implementation Complexity 🔄 | Resource Requirements ⚡ | Expected Outcomes 📊 | Ideal Use Cases 💡 | Key Advantages ⭐ |
|---|---|---|---|---|---|
| Age Range Specifications | Low–Moderate — simple DOB-based filters; boundary/ETL edge cases | Low — person & observation_period access; minimal compute | Demographic consistency; enables age-stratified analyses | Vaccine efficacy (18+), pediatric registries, geriatric cohorts | Improves relevance; reproducible cohorts; reduces age confounding |
| Diagnosis Code Requirements (ICD-10-CM, SNOMED CT) | Moderate–High — vocabulary traversal and hierarchy decisions | Moderate — vocab access, clinical curation, version management | Standardized, reproducible condition definitions across sites | Disease cohorts (T2D, HF, CKD); cross-site phenotyping | Captures clinical groupings; cross-vocabulary mapping; reduces manual lists |
| Laboratory Test Value Thresholds (LOINC) | Moderate — unit normalization and temporal logic required | Moderate — LOINC mapping, unit conversion tables, QC pipelines | Objective, high-fidelity phenotyping; guideline-aligned cohorts | HbA1c/eGFR/LDL thresholds for disease cohorts and trials | Quantifiable criteria; automatable checks; precise inclusion |
| Medication Exposure Windows (RxNorm) | Moderate — RxNorm/NDC mapping and exposure-window logic | Moderate–High — drug vocab, pharmacy data, mapping validation | Standardized drug exposure cohorts; supports safety and effectiveness studies | Statin/ACE inhibitor studies; pharmacovigilance; comparative effectiveness | Standardized drug definitions; ingredient hierarchies; washout support |
| Observation Period & Data Availability | Low–Moderate — inspect enrollment windows and continuity rules | Moderate — longitudinal records, completeness metrics | Sufficient baseline/follow-up; reduced selection bias; better outcome capture | Trials needing baseline, outcome studies, NLP validation | Ensures data completeness; improves causal inference validity |
| Procedure Code Inclusion (CPT, SNOMED CT) | Moderate — procedure mapping and modifier handling | Moderate — procedure vocab, billing/clinical verification | Precise intervention cohorts and device/procedure outcome measurement | Joint replacement registries, cardiac procedure studies, oncology interventions | Captures interventions accurately; supports surgical/outcome research |
| Visit Type & Healthcare Setting Specifications | Low–Moderate — map visit_concept/type ids; handle inconsistent codes | Low–Moderate — visit_occurrence, care_site metadata | Context-specific analyses; reduces setting-related bias | Inpatient safety, outpatient monitoring, telehealth studies | Ensures appropriate clinical context; enables setting comparisons |
| Exclusion Criteria Definition (Contraindications, Comorbidities) | High — complex temporal and multi-domain logic | High — multi-vocabulary queries, clinical expertise, extensive QC | Improved internal validity and patient safety; may reduce generalizability | Safety-driven trials (pregnancy, severe comorbidity exclusions); adverse event studies | Protects safety; reduces confounding; robust negative cohort definition |
From Criteria to Cohort: Your Key Takeaways for Robust Research
Throughout this article, we have dissected eight fundamental types of inclusion criteria, moving from theoretical concepts to practical, replicable code. The journey from a clinical question to a well-defined cohort is paved with meticulous planning and precise execution. Each inclusion criteria example we explored, from age ranges and diagnosis codes to specific lab values and medication exposures, underscores a central theme: the quality of your research is directly determined by the rigor of your cohort definition.
The transition from a research protocol written in prose to a functional, accurate query requires bridging the gap between clinical intent and data reality. This is where a standardized data model like OMOP and controlled vocabularies become indispensable. They provide the universal language needed to translate complex clinical ideas into unambiguous, computational logic that can be executed consistently across diverse datasets.
Mastering the Art and Science of Cohort Definition
Achieving robust and reproducible research is not just about writing code; it's about adopting a strategic mindset. Based on the examples and analyses provided, here are the most critical takeaways for your team:
- Document Everything: For every criterion, articulate the clinical rationale. Why was a specific diagnosis code chosen? What is the justification for the lab value threshold? This documentation is crucial for validation, peer review, and future replication.
- Embrace Iteration and Validation: Your first cohort definition is rarely your last. Collaborate closely with clinical experts to review initial cohort counts and patient profiles. Does the resulting group match clinical intuition? Sensitivity analyses, where you slightly alter criteria (e.g., changing an observation window by 30 days), are powerful tools for understanding the stability and sensitivity of your definition.
- Prioritize Specificity Over Simplicity: Vague criteria like "history of diabetes" are prone to misinterpretation. Instead, anchor your definitions in specific, measurable events: a diagnosis code from a physician encounter, a prescription for an anti-diabetic medication, or a qualifying lab result. This specificity is the bedrock of a strong inclusion criteria example.
Strategic Insight: The most common failure point in cohort definition is ambiguity. By using OMOP concept sets and documenting the logic behind each rule, you create an executable and auditable asset that is both scientifically sound and technically portable. This elevates your work from a one-off analysis to a reusable scientific building block.
Your Actionable Next Steps
To put these principles into practice, focus on integrating these habits into your workflow immediately:
- Standardize Your Vocabulary Management: Stop managing vocabulary files manually. Leverage a centralized terminology service to ensure your entire team is using the most current, correct concept identifiers for SNOMED CT, RxNorm, LOINC, and others. For more details, review the official documentation at docs.omophub.com.
- Codify Your Logic with SDKs: Transition from manual SQL queries to programmatic cohort definition using tools like the OMOPHub SDKs for Python and R. This makes your definitions more modular, easier to version control, and significantly faster to deploy.
- Implement Peer Review for Cohorts: Treat your cohort definitions like production code. Institute a peer-review process where another researcher or data scientist validates the logic, checks the concept sets, and questions the assumptions before the analysis begins.
By internalizing these lessons and adopting these practices, you transform the complex task of cohort definition from a research bottleneck into a strategic advantage. You will not only accelerate your study timelines but also produce more reliable, transparent, and impactful results that can truly advance clinical understanding.
Ready to eliminate the complexity of vocabulary management and accelerate your research? OMOPHub provides a turn-key, enterprise-grade terminology service and powerful SDKs that allow you to implement any inclusion criteria example with speed and precision. Visit OMOPHub to see how you can build better cohorts, faster.