A Practical Guide to the Healthcare Common Data Model
A common data model (CDM) is essentially a universal translator for messy, disconnected information. In healthcare, this means taking all the complex data from countless sources—electronic health records (EHRs), insurance claims, lab systems, you name it—and reshaping it into a single, consistent format.
The goal is to get different systems to "speak the same language." When they do, it opens the door for large-scale, reproducible research and the kind of advanced analytics that was once impossible.
Why a Common Data Model Is Essential in Healthcare
Imagine you’re trying to build a house with bricks from a dozen different suppliers. Each one has a slightly different size, shape, and material. You’d spend more time cutting, sanding, and forcing pieces to fit than you would actually building. That’s a perfect picture of healthcare data analytics without a common data model.
Data comes at us in a thousand different formats. One hospital might record a diagnosis using a standard ICD-10 code, while another uses its own internal coding system, and a third just has a doctor's free-text notes. It’s chaos. A CDM, and specifically the Observational Medical Outcomes Partnership (OMOP) model, brings order to that chaos by providing a shared, predefined blueprint for the data.
A Common Data Model isn't just about organizing data; it's about creating a shared understanding. It establishes a consistent structure and vocabulary that makes large-scale, collaborative research possible across different organizations and even countries.
Before we dive deeper into OMOP's structure, let's look at the core components that make it work. The model is built on two main pillars: the standardized data tables that hold clinical information and the standardized vocabularies that give that information meaning.
Here’s a quick breakdown of how those pieces fit together:
Key Components of the OMOP Common Data Model
| Component | Function |
|---|---|
| Standardized Data Tables | These tables (e.g., PERSON, CONDITION_OCCURRENCE, DRUG_EXPOSURE) store the actual clinical data in a predefined structure. Every patient journey is mapped into these consistent columns and rows. |
| Standardized Vocabularies | This is the "dictionary" that translates all the different source codes (like ICD-10, SNOMED, RxNorm) into a single, standard set of concepts. It ensures "Type 2 Diabetes" means the same thing everywhere. |
This combination of a fixed structure and a controlled vocabulary is what gives the OMOP CDM its power.
The Universal Adapter for Health Data
The OMOP CDM is a lot like a universal power adapter you'd pack for an international trip. Your US-made laptop can't plug directly into a European outlet, but the adapter makes it work seamlessly. In the same way, OMOP lets researchers plug their analytical tools into datasets from all over the world, knowing the underlying structure is always the same.
This standardization is a game-changer for a few key reasons:
- Enabling Large-Scale Research: It lets researchers pool data from multiple hospitals or even countries to get the statistical power they need to make meaningful discoveries.
- Improving Reproducibility: When everyone works from the same data structure, studies are far easier for other teams to validate and reproduce. The "it worked on my machine" problem fades away.
- Accelerating Analytics: Data scientists can build an analysis pipeline once and run it across any OMOP-compliant database without a major overhaul. This is a massive time-saver for projects in fields like health economics and outcomes research, which you can explore in our HEOR use case overview.
Practical Application Using OMOPHub SDKs
Getting your source data into the OMOP CDM format often involves the tricky step of mapping local, proprietary codes to the standard OMOP vocabularies. This used to be a huge headache.
Thankfully, tools like the OMOPHub SDKs for Python and R make this much simpler. For instance, a developer can use a single function to look up the standard concept for a local diagnosis code without having to download and manage massive vocabulary files.
Here’s a quick conceptual example of what that looks like with the Python SDK:
# Import the OMOPHub client
from omophub.client import Client
# Initialize the client with your API key
# You can also set the OMOPHUB_API_KEY environment variable
client = Client(api_key="YOUR_API_KEY")
# Search for a standard concept related to 'diabetes'
# This is useful for exploratory analysis.
# See https://docs.omophub.com/sdks/python/vocabulary#search_concepts for more options.
concepts = client.vocabulary.search_concepts(query="diabetes", standard_concept="Standard")
# Print the first result
if concepts:
print(concepts[0])
else:
print("No concepts found.")
A simple command like this abstracts away immense complexity. It frees up developers to focus on the logic of their data transformation pipelines, not the logistics of vocabulary management. By bringing this level of standardization to otherwise disconnected data, the common data model provides the very foundation for the future of evidence-based medicine.
How the OMOP CDM Structures Patient Data
The real magic of the OMOP common data model is its person-centric design. It’s not just a database; it’s a framework for rebuilding a patient's entire health journey into a story that can be analyzed. Think of it as meticulously assembling a detailed puzzle where every piece is a specific clinical event in that person’s life.
To really get a handle on it, you can split the core tables into two groups: those that set the scene and those that capture the action. The first set, what we call the Health System Data tables, establishes the "who," "where," and "when" of patient care.
These are the foundational pieces:
- PERSON: This is the anchor. Every row is a unique individual, holding basic demographic info like their birth year and gender.
- VISIT_OCCURRENCE: This table tracks every single time a person interacts with the healthcare system, whether it’s a quick outpatient visit or a long hospital stay.
- PROVIDER and CARE_SITE: These tables give you the crucial context of which doctors, nurses, clinics, and hospitals were involved.
Capturing the Clinical Story
With that foundation in place, the Clinical Event tables step in. This is where the nitty-gritty details of a patient’s health narrative get recorded. Each table is built to hold a specific type of clinical data, effectively creating a detailed log of every diagnosis, treatment, and observation over time.
For example, the model has specific tables for things like:
- CONDITION_OCCURRENCE: For every diagnosed condition, from a common cold to a chronic illness.
- DRUG_EXPOSURE: This logs every medication prescribed, taken, or administered.
- PROCEDURE_OCCURRENCE: Documents any and all medical procedures a patient undergoes.
- MEASUREMENT and OBSERVATION: These are catch-alls for lab results, vital signs, and other important clinical findings.
By linking every clinical event back to a specific person and a specific visit, OMOP creates an incredibly rich, longitudinal record. This is what lets researchers ask tough questions like, "How many patients with Type 2 Diabetes were prescribed Metformin during an inpatient stay and later developed kidney disease?"
This structure turns a mountain of disconnected data points into a cohesive history you can actually query. The diagram below does a great job of showing how all that messy source data gets funneled through the common data model to produce standardized, analysis-ready information.
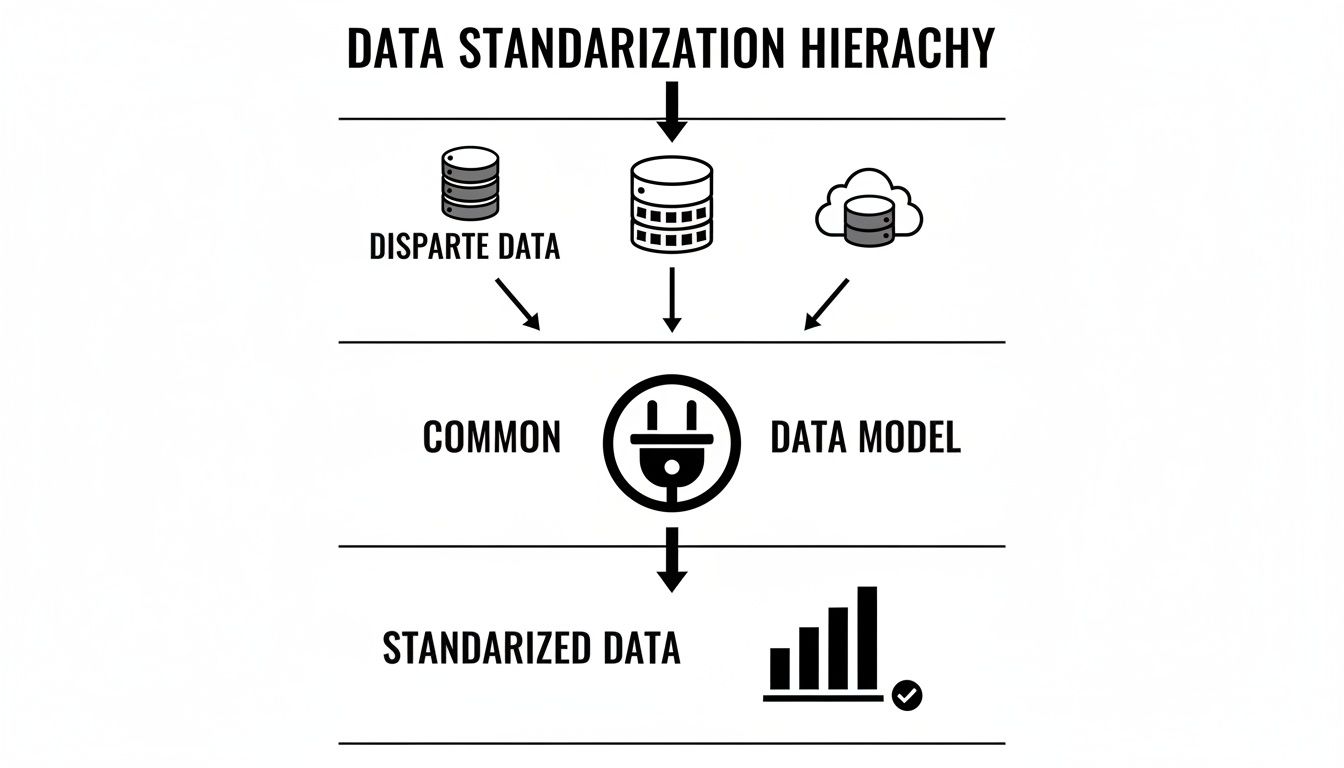
As you can see, the CDM acts as a critical translator, taking all those different inputs and shaping them into a consistent format that makes reliable analytics possible.
Unlocking Comparative Research
This ability to standardize and connect data is what makes modern healthcare analytics, especially comparative effectiveness research, possible. Take, for instance, a study that involved two very different US health systems—one civilian, one federal. They mapped their EHR data to the OMOP CDM.
Despite huge underlying differences—the federal dataset had an average of 6.44 outpatient visits per person-month versus just 2.05 in the civilian system—the model harmonized it all. Even though one system tracked medication orders and the other tracked medication fills, OMOP unified them. This allowed for consistent analysis across both datasets. For a closer look at the methodology, you can read the full research on normalizing these data disparities.
This unified structure is a game-changer. It means that analytical code written for one OMOP database can run on any other OMOP database without modification. A simple query to count patients with a specific condition can be executed across dozens of institutions. Trying to do that without a common data model would be an absolute nightmare.
Ultimately, the real value here isn't just about clean data storage. It's about creating a launchpad for discovery.
Mastering OMOP Vocabularies for Data Harmony
A standardized structure for patient data is a great start, but it only gets you halfway there. The real magic of the OMOP common data model, the part that transforms it from a tidy database into a powerhouse for clinical insight, is its standardized vocabularies.
Think of it like this. The table structure tells you where to put the data—you have a jar labeled "flour," another for "sugar," and so on. But the vocabularies make sure we all agree on what "flour" actually is. Without that shared definition, one person might fill the jar with whole wheat flour while another uses almond flour. Any recipe you try to create from that pantry is going to be wildly inconsistent.
This is the core challenge of semantic interoperability. It’s not enough to know a column holds "drug codes." You have to know that one hospital's internal code 40213220 and another's code 243681002 both point to the exact same thing: "Lisinopril 10 MG Oral Tablet." This is the level of precision OMOP achieves by mapping thousands of local, source-specific codes to a single, unified set of standard concepts.

The Role of Standard Terminologies
The OMOP CDM pulls in several globally recognized terminologies to build out its vocabulary system. Each one is a specialist, covering a different domain of healthcare data.
- SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms): This is the heavyweight for clinical findings, conditions, and procedures. Its deep hierarchy is a researcher's dream, letting you query for something broad like "any malignant neoplasm" or drill down to a highly specific diagnosis.
- RxNorm: This is the master translator for all things pharmacy. It untangles the web of branded drugs, generics, active ingredients, and dose forms, ensuring that "Tylenol" and a generic "Acetaminophen 500mg tablet" are correctly analyzed as the same active substance.
- LOINC (Logical Observation Identifiers Names and Codes): When it comes to lab tests and clinical measurements, LOINC is the gold standard. It provides a universal code for every test imaginable, from a simple blood glucose reading to a complex genetic panel.
By mapping all the messy, disparate source data to these standard concepts, OMOP creates true data harmony. This is what makes powerful, reproducible research possible across completely different datasets.
The Developer Challenge and a Modern Solution
Here’s the catch: managing these vocabularies is a huge technical lift. The complete OHDSI ATHENA vocabulary dataset is enormous, often ballooning to over 50GB. Just hosting and querying it requires a beefy database server. And on top of that, you have to keep it updated with regular releases, adding yet another operational headache for your data engineering team.
This infrastructure burden has historically been a major barrier to OMOP adoption. Teams spend valuable time and resources on database administration instead of focusing on what really matters: building data pipelines and generating insights.
This is exactly the problem that developer-first platforms like OMOPHub were created to solve. By offering instant REST API and SDK access to the complete, up-to-date ATHENA vocabularies, OMOPHub completely eliminates the need to host anything yourself. Developers can just install an SDK, drop in an API key, and start mapping concepts programmatically in minutes.
A Practical Code Example in Python
Let’s see what this looks like in the real world. Imagine an ETL developer needs to map a source condition code for "Type II Diabetes Mellitus" to its standard OMOP concept. With the OMOPHub Python SDK, this once-clunky task becomes a simple API call.
# Import the OMOPHub client
from omophub import Client
# Initialize the client with your API key
# The client will automatically use the OMOPHUB_API_KEY environment variable if not provided
client = Client()
# Define the search query
# We'll specify the source vocabulary and code, then ask for its standard equivalent
# In this case, 'ICD10CM' is the source vocabulary
# and 'E11.9' is the code for Type 2 diabetes without complications
# For more info, see: https://docs.omophub.com/sdks/python/vocabulary#get_concept_maps
concept_maps = client.vocabulary.get_concept_maps(
source_vocabulary_id="ICD10CM",
source_concept_code="E11.9"
)
# The result is a list of mappings; we'll take the first one
if concept_maps:
standard_concept = concept_maps[0].concept
print(f"Source Code: 'E11.9' maps to Standard Concept ID: {standard_concept.concept_id}")
print(f"Standard Concept Name: {standard_concept.concept_name}")
else:
print("No standard concept mapping found.")
This little snippet shows just how easily a developer can find the correct standard concept_id they need to populate the CONDITION_CONCEPT_ID field in the CONDITION_OCCURRENCE table. No database lookups, no local CSV files. You can dig into the full range of possibilities in the official OMOPHub documentation.
This API-driven approach not only saves an incredible amount of time but also enforces consistency and accuracy by ensuring you're always referencing the latest vocabulary versions. You can read more about why this matters in our guide to vocabulary versioning.
There’s a reason the OMOP common data model has seen such explosive global adoption: this combination of a solid structure and harmonized vocabularies just works. By 2020, more than 330 databases across 30+ countries had been converted to this standard, which helped double the number of published studies from the previous year. That surge shows how powerful this model is for large-scale research—a trend that's only picking up speed as tools continue to simplify the tricky parts like vocabulary integration. To learn more, check out the full research about OMOP's global adoption.
A Practical Guide to Your ETL and Data Mapping Strategy
Once you've got your head around the OMOP common data model and its vocabularies, the real work begins: building the pipeline that gets your data into it. This is where the Extract, Transform, and Load (ETL) process comes into play. Think of it as the engine that takes raw, messy source data and turns it into a clean, standardized OMOP database.
Forget about a single "big bang" conversion. A truly effective ETL strategy is an iterative, thoughtful process of mapping, testing, and refining.
The whole journey kicks off with a deep dive into your source data. Before you write a single line of code, you have to understand the landscape you're working with. What data do you have? Where does it live? How is it coded? This initial exploration is non-negotiable—it's where you'll spot the landmines like inconsistent data entry, fields full of missing values, or obscure local coding systems that will trip you up later.
Core Principles for Successful Data Mapping
With a clear picture of your source data, you can start the mapping process. This is the heart of the "Transform" step, where you write the rules that translate a source record into its new home in an OMOP table. A disciplined, piece-by-piece approach is your best friend here.
It's best to start with the foundational tables and build out from there:
- Map Person and Visit Data First: Get your patient demographics into the
PERSONtable and their healthcare encounters into theVISIT_OCCURRENCEtable. These two tables provide the bedrock context for every other clinical event. - Tackle Clinical Domains Iteratively: Next, move on to the clinical event tables like
CONDITION_OCCURRENCEorDRUG_EXPOSURE, but do them one at a time. Trying to map everything at once is a surefire recipe for chaos and bugs. - Validate at Each Step: After mapping each domain, stop and run data quality checks. Do the record counts look right? Are the
concept_idfields populated correctly? This constant feedback loop helps you catch issues early, when they're much easier to fix.
The goal of ETL isn't just to move data; it's to preserve and enhance its meaning. A successful mapping strategy ensures that a diagnosis code from a source system retains its precise clinical intent when it becomes a standard concept in the OMOP CDM.
You'll also need clear, documented rules for handling common hurdles like missing data. For instance, if a source end_date for a condition is null, your ETL logic should consistently default to using the start_date to maintain data integrity across the board.
Streamlining Vocabulary Mapping with the R SDK
Let’s be honest: one of the most tedious parts of any OMOP ETL project is mapping all your local source codes to the standard OMOP vocabulary concepts. This process is a prime candidate for automation. Using tools like the OMOPHub SDKs can slash the manual effort involved and let developers programmatically find the correct standard concepts.
For example, a developer using the OMOPHub R SDK can efficiently map a long list of local drug codes to their standard RxNorm equivalents. This doesn't just speed up development time; it also makes sure the mappings are consistent and based on the latest vocabulary release.
Here’s a quick look at how you might use it in an R script to find standard concepts for a list of source codes.
# First, ensure you have the omophub client installed
# install.packages("omophub")
# Load the library
library(omophub)
# Initialize the client. It will use the OMOPHUB_API_KEY environment variable.
client <- Client$new()
# Imagine we have a list of source ICD-10-CM codes we need to map
source_codes <- c("I10", "E11.9", "J45.909")
# Use the get_concept_maps_bulk function for efficient batch processing
# For more info, see: https://docs.omophub.com/sdks/r/vocabulary#get-concept-maps-bulk
concept_maps <- client$vocabulary$get_concept_maps_bulk(
source_vocabulary_id = "ICD10CM",
source_concept_codes = source_codes
)
# Print the results to see the standard concept mappings
print(concept_maps)
This simple script automates what would otherwise be a soul-crushing task of manual lookups or writing complex database joins. For a deeper dive into all the functions, the official OMOPHub documentation has plenty of examples. And while this example is in R, the same kind of functionality is available in other languages—check out our recent Python SDK release notes for more.
By adopting these field-tested practices and modern tools, your team can navigate the complexities of data mapping with far greater efficiency and accuracy. This approach turns the ETL process from a major bottleneck into a systematic, scalable part of your common data model implementation.
How OMOP Stacks Up Against Other Data Models
Picking a common data model is a foundational decision, and the OMOP CDM isn't the only game in town. It sits in a landscape of other well-established standards, so making the right choice means understanding how it differs from major players like PCORnet, i2b2, and Sentinel.
The goal here isn’t to crown a single winner. It's to appreciate the different design philosophies that drive each one.
Each of these models was created to solve a specific problem, and that origin story still shapes its structure and best-fit use cases today.
Design Philosophies and Core Strengths
The OMOP CDM was built from the ground up for large-scale observational research. Its whole purpose is to achieve deep semantic consistency across wildly different datasets. The secret sauce is its granular, person-centric table structure combined with that incredibly rigorous system of standardized vocabularies.
This design makes it the go-to choice for federated network studies, where the exact same analysis code needs to run reliably across dozens of separate databases to generate reproducible evidence.
Other models, however, were optimized for different tasks:
-
PCORnet CDM: Born out of the Patient-Centered Outcomes Research Network, this model is tailored for pragmatic clinical trials and comparative effectiveness studies. Its structure often mirrors the source EHR data more closely, which can sometimes make the initial ETL process a bit more straightforward.
-
i2b2 (Informatics for Integrating Biology and the Bedside): This model is all about speed. It uses a "star schema" database design that’s incredibly efficient at cohort discovery. If your main question is, "How many patients in our hospital have these specific characteristics?" i2b2 is built to give you that answer, fast.
-
Sentinel CDM: Created for the FDA’s Sentinel Initiative, this model has a very specific job: post-market drug safety surveillance. It’s intentionally rigid and less flexible than OMOP, a feature designed to ensure absolute consistency for its critical regulatory mission.
The core trade-off usually boils down to upfront effort versus downstream analytical power. OMOP demands a significant investment in vocabulary mapping, but that work unlocks a universe of standardized, cross-platform analytics. Other models might get you converted faster for more targeted needs.
A Side-by-Side Look
When you get down to the details, the different strengths and weaknesses of each data model become much clearer. The table below breaks down the key distinctions between the four major healthcare CDMs.
Comparison of Major Healthcare Common Data Models
| Feature | OMOP CDM | PCORnet CDM | i2b2 | Sentinel CDM |
|---|---|---|---|---|
| Primary Goal | Large-scale observational research & evidence generation | Pragmatic clinical trials & comparative effectiveness | Rapid cohort discovery & feasibility analysis | Post-market drug safety surveillance |
| Data Structure | Person-centric, highly normalized relational model | Closer to source EHR/claims structure; less normalized | EAV/CR (star schema) optimized for querying counts | Highly focused, less flexible; purpose-built for safety |
| Vocabulary | Centralized, mandatory, and rigorously standardized | Uses a mix of standard and site-specific codes | Primarily local ontologies; can be federated | Tightly controlled, specific set of standard codes |
| Flexibility | Highly extensible and adaptable for new domains | Moderately flexible; focused on clinical trial data | Flexible for local institutional needs | Very rigid and purpose-built by design |
| Community | Large, active, and global open-source community (OHDSI) | Network-driven (PCORnet); strong US presence | Academic and institutional user base | Government-led (FDA initiative) |
As the comparison shows, the choice isn't about which model is "best" in a vacuum, but which is best suited for your specific analytical goals, technical resources, and long-term data strategy.
In comprehensive evaluations, standardization through Common Data Models like OMOP consistently comes out on top. In one recent analysis, it scored a perfect 21+ across key criteria like suitability, popularity, adaptability, and interoperability—ahead of PCORnet (20+), i2b2 (17+), and Sentinel (16+). This is largely thanks to its open-source nature, robust versioning, and proven success in federated learning. You can dig into the full comparative research on data model suitability to see the detailed breakdown.
For development teams working with EHR data, OMOP’s relational tables for concepts and relationships are a natural fit for integration tools. This is precisely why platforms like OMOPHub, with its dedicated SDKs, can simplify the otherwise complex process of vocabulary management and integration.
By understanding this landscape, you can confidently choose the model that aligns with your organization's vision for turning data into a powerful, future-proof asset.
Got Questions About the Common Data Model? We've Got Answers.
Moving your data into a common data model like OMOP is a major undertaking. It’s totally normal to have a long list of questions as you shift from planning to actually doing the work. Let's tackle some of the most common ones that pop up when teams get their hands dirty with a real implementation.
How Long Does a Typical OMOP Data Mapping Project Take?
This is the big one, and the honest answer is: it depends. The timeline for an OMOP Extract, Transform, and Load (ETL) project can swing wildly based on just how complex and clean your source data is, not to mention your team's familiarity with the model.
For a small, well-structured dataset from a single source, you might get it done in a few weeks. But if you're wrestling with a sprawling Electronic Health Record (EHR) system holding decades of messy data and a tangled web of local codes, you should probably brace for a 6 to 12-month journey, or possibly even longer.
The timeline really boils down to a few key factors:
- Source Complexity: How many source tables are you dealing with, and how neatly do they fit into OMOP’s structure?
- Data Quality: Are you swimming in missing values, inconsistent formats, or free-text fields that need to be wrangled into submission?
- Vocabulary Mapping: How many different coding systems (like ICD-9, ICD-10, and internal proprietary codes) need to be mapped over to standard OMOP concepts? This is often the heaviest lift.
- Clinical Validation: How much time will you need for clinical experts to review the mapping logic? This step is critical for ensuring the final data is scientifically sound.
This is where modern tooling can really change the game. For instance, tackling the vocabulary mapping bottleneck with a platform like OMOPHub can shave a significant amount of time off your project. By giving you immediate API access to standardized terminologies, you completely sidestep the headache of setting up, hosting, and maintaining your own vocabulary database—a task that can easily burn weeks of your team's time.
What Are the Biggest Challenges of Managing OMOP Vocabularies?
The standardized vocabularies are the lifeblood of the OMOP CDM, but they come with two major hurdles: infrastructure and expertise.
First off, the infrastructure required is no small thing. The full OHDSI ATHENA vocabulary set is massive—often over 50GB—and demands a pretty beefy database server just to host it and run queries effectively. On top of that, these vocabularies aren't static. They get updated regularly, which means your data engineering team is on the hook for an ongoing operational burden just to keep your local copy current.
Second, you need some serious expertise to do the mapping correctly. It’s not just about finding a code that looks right; it’s about choosing the one standard concept that truly captures the original clinical meaning. This often requires someone who speaks fluent SNOMED CT, RxNorm, and LOINC and can navigate the subtle but critical differences between terms.
This is precisely the problem platforms like OMOPHub were built to solve. They handle all the heavy lifting on the infrastructure side and offer powerful API endpoints for searching and mapping, making life dramatically easier for ETL developers and researchers.
If you want to see what this looks like in practice, check out the OMOPHub Python SDK documentation. You'll see how a once-complex lookup becomes a simple, clean function call.
Can I Extend the OMOP CDM with Custom Tables or Columns?
Absolutely. The OMOP common data model was designed with the understanding that real-world data is messy and that not every piece of information will have a perfect home in the standard tables. The model is built to be extended.
The official documentation gives clear guidance on adding your own custom tables or columns. The trick is to do it in a way that doesn't break compatibility with standard tools. A common best practice is to keep your extensions documented and physically separate—for example, by creating new tables with a special prefix like x_custom_table.
This approach gives you the best of both worlds:
- It Preserves Compatibility: Standard OHDSI analytics tools can still run perfectly on the core, standardized part of your database.
- It Maintains Clarity: Everyone knows exactly what's part of the universal standard and what's a custom addition for your organization.
This way, you can accommodate your unique research questions without giving up the incredible power that comes from standardization.
How Does the OMOP CDM Address Data Privacy and HIPAA Compliance?
This is a critical point of clarification: the OMOP CDM is a data structure, not a security tool. It doesn't have any built-in security controls. Responsibility for data privacy, HIPAA compliance, and GDPR adherence rests entirely with the organization implementing the model.
Making your OMOP instance compliant requires a comprehensive security strategy that your team puts in place. This almost always includes:
- De-identification: Stripping out or masking Protected Health Information (PHI) before or during the ETL.
- Access Controls: Setting up strict, role-based permissions for who can access the database.
- Encryption: Making sure data is encrypted both at rest (on the disk) and in transit (across the network).
- Auditing: Keeping detailed logs of who accessed what data, and when.
When you bring third-party services into your workflow, you have to make sure they meet your security standards. A service that handles vocabulary lookups, for instance, should offer end-to-end encryption and adhere to enterprise-grade security practices. This ensures your entire data pipeline, from end to end, is buttoned up. You can see an example by reviewing the OMOPHub security documentation.
Ready to eliminate the biggest bottleneck in your OMOP ETL pipeline? OMOPHub provides instant API and SDK access to the complete, always-updated OHDSI ATHENA vocabularies, so your team can focus on building data pipelines, not managing databases. Get started in minutes at https://omophub.com.


